













| General Information: |
|
| Name(s) found: |
pst3 /
SPBC1734.16c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1154 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC Rpd3L-Expanded complex [IDA cytosol [IDA Rpd3S complex [IDA |
| Biological Process: |
chromatin remodeling
[NAS]
regulation of transcription, DNA-dependent [IC chromatin silencing [ISS histone deacetylation [IC double-strand break repair via nonhomologous end joining [ISS] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVMNVPVDS ERDNPGDKVE TQSDKNHLPK ASPSQSQSPV NTSLHNGDGK DNGVATEPVE 60
61 NKQILSERSV TRDDYEKGKT IVSSLALSSI SGKDGSISSQ NAEGLSSSSN RPLDVNDALS 120
121 YLELVKYYFS ERREIYNRFL EIMRDFKSQA LDTLGVINRV SELFNGYPQL IEGFNTFLPS 180
181 GYKIEVQLDS SNTSVVRVGT PMHPLPQQGV QSTLPVAPSN EDQRTMESTS PTDSQPQPSA 240
241 PNLVSSTENE KPRVDFNYAI AYMNKVKARY PPNSDTYMEF LGVLRTYQKA QKSIFEVRAR 300
301 VAEIFKDSPD LLEEFKLFLP DNVDSTEPST PNVQKSPNRL PPVGNFSLPP SAPVREKRNR 360
361 PAHSAQISRS ISKTSRMYRQ TAEEPLNSYS LHVYPQKITA PTSPYAATQE ELLAFTTIRQ 420
421 HLPDTLAFHK FLELLHLYRE KLLDKTELLN SFSKLVRNDN LTLWFSEFIR WSDNPILVKN 480
481 EPVDERVYLP ETFECISLTY RKLPDSWKQD KCSGRDDLDN SVLNDDYISV APKPSHVKNI 540
541 MHHENQYLQA LQLVEDERYD YDRVLNTTES AIKILANFCE PTIHEHLETA LQELERSKRI 600
601 IKNALIIVYG KEHANLALDT LFKKLPTAAP VLLKRIKTKD QEWRRSKREW SKIWRQIEKK 660
661 NAQAAFDDRY CRIEGRDRRG LSYSRILRDI DDIYQRQKHR IDGAKLGFQF TQVLCDSLIF 720
721 LNILRLSDAQ LTNSSFYSYA DKGRISAVLK ALLSQFFGIP LPREALETNL ASENIESVKK 780
781 HRDGLSKIFI RPESADNSNN TNVSFQTDET QTEDETMSDI HPDDVENHSK SKFLGEESKN 840
841 IIGYNFFGNA TMYVLFRLIC VCYSRLEHIK LFVESSTIYA SSTGGYENIL NICEKYLKGS 900
901 CSRLEFRKYL QKFNNETCYM ICSIERLLKV IFYRIHEILL DPKLGQLLLL FESDGANSVT 960
961 TPREQMVYRN HVESILAPES KIFNMRWYPL EKRLCIQQLL PADLTMHDFE NPAKAFMYYV1020
1021 DSYAISHITE GVDLMQVKMP FLRRSLQRIS QQGYLAGRGS GRLHSLFNEH FCKSNLQLFF1080
1081 STDTYVIFFE PNTENVYINS YNLWVDQSSQ SKKQNRTTNW RRWLESDEGW RKSKANTDIK1140
1141 FFSETTLDQC IEAM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
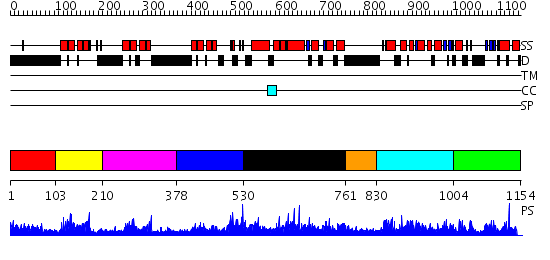
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..102] | 106.155997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [103..209] | 30.0 | Solution structure of the NRSF/REST-mSin3B PAH1 complex | |
| 3 | View Details | [210..377] | 22.221849 | No description for 2f05A was found. | |
| 4 | View Details | [378..529] | 2.5 | No description for 2f05A was found. | |
| 5 | View Details | [530..760] | 1.076967 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [761..829] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [830..1003] | 1.051981 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1004..1154] | 1.044979 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)