













| General Information: |
|
| Name(s) found: |
hsk1 /
SPBC776.12c
[Sanger Pombe]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 507 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Dbf4-dependent protein kinase complex [TAS |
| Biological Process: |
DNA replication initiation
[IGI DNA replication checkpoint [IGI replication fork protection [IGI chromosome organization [IMP G1/S transition of mitotic cell cycle [IMP replication fork arrest [IGI protein amino acid phosphorylation [IEA] mating type switching [IMP regulation of centromeric sister chromatid cohesion [IGI |
| Molecular Function: |
ATP binding
[IDA protein binding [IPI protein heterodimerization activity [IPI protein serine/threonine kinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEAHITLSP KVTHEQQTDI DSECEITEVD DENVNENKSQ EMIQDIPARD REEIENITRT 60
61 FVELQENYRL IEKIGEGTFS SVYKAEDLHY GRYINDWDIQ SEVLKESSFG KEKIPVNEDS 120
121 RKPKYVAIKK IYATSSPARI YNELEILYLL RGSSVIAPLI TALRNEDQVL VVLPYYEHTD 180
181 FRQYYSTFSY RDMSIYFRCL FQAMQQTQTL GIIHRDIKPS NFLFDVRTKH GVLVDFGLAE 240
241 RYDGRQQSHS CRCTNSNAAE LAHDFSIAQE TSLGYIKNDT RPSKRANRAG TRGFRAPEVL 300
301 FKCSSQSPKV DIWSAGVILL SFLTKRFPMF NSKDDVDALM EIACIFGKSE MRQCAALHGC 360
361 TFETNVSTLT EKRVNFRKLI LWASCGSASI YKEKLRHKPS QEERLCLDFL EKCLELDCNK 420
421 RISAEEALDH DFLYLDNLAY EKKDDDTAFD NSFGETSFEK DEDLTAKHLS HILDFKEQEE 480
481 TDEPTSLSKR KRSIDEILPN DALQDGA |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
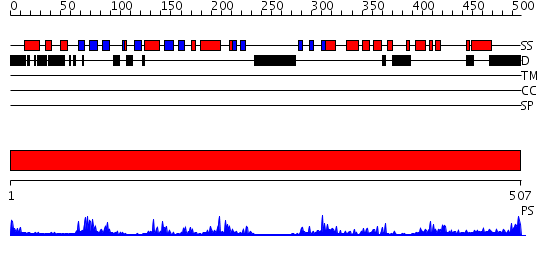
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..507] | 53.0 | MAP kinase p38-gamma |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)