













| General Information: |
|
| Name(s) found: |
FBpp0292319
[FlyBase]
ttk-PA / FBpp0085190 [FlyBase] ttk-PB / FBpp0085188 [FlyBase] ttk-PE / FBpp0085186 [FlyBase] |
| Description(s) found:
Found 55 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 813 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
polytene chromosome [IDA] |
| Biological Process: |
regulation of cell shape
[IMP]
R7 cell development [TAS] peripheral nervous system development [TAS] dorsal appendage formation [IMP] brain morphogenesis [IMP] neuron development [IMP] inter-male aggressive behavior [IMP] regulation of transcription from RNA polymerase II promoter [NAS] dendrite morphogenesis [IMP] negative regulation of transcription from RNA polymerase II promoter [NAS] locomotion involved in locomotory behavior [IMP] negative regulation of transcription, DNA-dependent [IDA] startle response [IMP] ovarian follicle cell development [IMP] cell fate determination [TAS] |
| Molecular Function: |
DNA binding
[IEA]
zinc ion binding [IEA] protein homodimerization activity [IPI] protein binding [IPI] specific RNA polymerase II transcription factor activity [NAS] chromatin binding [IDA] transcription repressor activity [NAS] specific transcriptional repressor activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKMASQRFCL RWNNHQSNLL SVFDQLLHAE TFTDVTLAVE GQHLKAHKMV LSACSPYFNT 60
61 LFVSHPEKHP IVILKDVPYS DMKSLLDFMY RGEVSVDQER LTAFLRVAES LRIKGLTEVN 120
121 DDKPSPAAAA AGAGATGSES TATTPQLQRI QPYLVPQRNR SQAGGLLASA ANAGNTPTLP 180
181 VQPSLLSSAL MPKRKRGRPR KLSGSSNGTG NDYDDFDREN MMNDSSDLGN GKMCNESYSG 240
241 NDDGSDDNQP NAGHTDDLNE SRDSLPSKRS KNSKDHRVVS HHEDNSTSVT PTKATPELSQ 300
301 RLFGSSSTTI SATAPGGSST GPSETISLLE ISDERESAPV HLPTILGLKI RAINTTTPAQ 360
361 QGSPQTPTKS KPKIRQATGS NNSNSLLKQQ LRGGAKDPEV PPATRITGAV TPNAALNAEE 420
421 QSKEMPKKNQ DEVNACIGLH SLANAAEQQA AQVASTGNLH HQLLLHMAAN NSMLNTTDYY 480
481 QQQQQESPSS AGQFMDDDLE LLSLNDQQDK SDEPDHEMVT LADENAGLPG YQGNEAEATP 540
541 AQEDSPAAET ATAPPPAPRS GKKGAKRPIQ RRRVRRKAQS TLDDQAEHLT EMSVRGLDLF 600
601 RYASVVEGVY RCTECAKENM QKTFKNKYSF QRHAFLYHEG KHRKVFPCPV CSKEFSRPDK 660
661 MKNHLKMTHE NFTPPKDIGA FSPLKYLISA AAAGDMHATI YQQQQDHYHR QLAEQLEQQN 720
721 ASFDSRDSSL ILPDVKMEHA EDQDAEQEAE LSDGGYDASN PAAAAAAMLS LQQDVIIKDE 780
781 IQISPSPSPT PPASCAVAEG KSLALASTAQ TAT |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
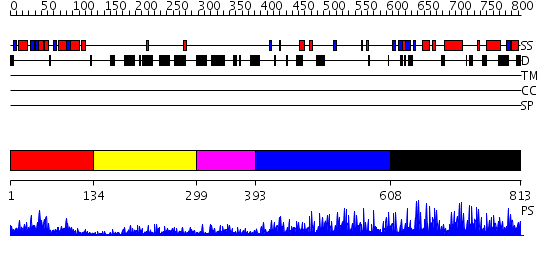
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 23.0 | Crystal Structure of the B-Cell Lymphoma 6 (BCL6) BTB domain to 2.2 Angstrom | |
| 2 | View Details | [134..298] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [299..392] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [393..607] | 37.0 | No description for 2i13A was found. | |
| 5 | View Details | [608..813] | 10.69897 | Five-finger GLI1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)