













| General Information: |
|
| Name(s) found: |
Apc2-PA /
FBpp0083937
[FlyBase]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1067 amino acids |
Gene Ontology: |
|
| Cellular Component: |
apicolateral plasma membrane
[TAS]
nucleus [TAS] dendrite [IDA] adherens junction [TAS] cytoplasm [TAS] cleavage furrow [IDA] cell cortex [IDA] apical cortex [IDA] protein complex [IPI] neuronal cell body [IDA] actin filament [IDA] microtubule associated complex [ISS] |
| Biological Process: |
cell-cell adhesion
[IGI]
establishment of mitotic spindle localization [TAS] chitin-based larval cuticle pattern formation [IGI] chitin-based embryonic cuticle biosynthetic process [IMP] head involution [IGI] syncytial nuclear migration [IMP] cytokinesis [TAS] axon guidance [IGI] cell adhesion [IMP][NAS] positive regulation of proteolysis [TAS] brain development [IGI] imaginal disc pattern formation [IGI] microtubule-based process [ISS][TAS] neuroblast proliferation [IGI] medulla oblongata development [IGI] negative regulation of transcription, DNA-dependent [TAS] Wnt receptor signaling pathway [TAS] negative regulation of Wnt receptor signaling pathway [IGI] regulation of Wnt receptor signaling pathway [IGI][IMP] pseudocleavage during syncytial blastoderm formation [IMP] oocyte localization during germarium-derived egg chamber formation [IGI][IMP] |
| Molecular Function: |
protein binding
[IPI]
microtubule binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLNESDATR LELTRNFLEL SRNPETCTAL RSSDCIQLLV QILHANDEGL STAKKYASQA 60
61 LHNIVHNNPE EKERQREVKM LRLLDQILDY CNFLHTQLQS GGEAIADDED RHPLAAMKLL 120
121 MKASFDEEHR QTMCELGALK AIPNLVHLDH AVHGPAAGRE QCNALRSYGL MALTNLTFGD 180
181 ENVHNKSYLC GQRQFMEVVI AQLNTAPDEL LQVLAGVLRN LSWRADKHMK TIFNELGTVT 240
241 SLARAAMQNK NENTLKAILS ALWNLSAHCS TNKAEFCAVD GALAFLVGML SYEGPSKTLK 300
301 IIENAGGILR NVSSHIAVCE PYRQILRRYN CLAILLQQLK SESLTVVSNS CGTLWNLSAR 360
361 CPEDQQYLID HNAIPLLRAL ISSKNSMIAE GSASALKNLV NFRATQELMP NGDGGSLPLD 420
421 KEAGHGGTLP RRFSSLRLSS NPTGSLKKVR PSTVSTTGFL NRKCESRESI YSGKSDSTKY 480
481 STKSEGAKNP FEIVTPTEEQ PIDYSMKYME HKPNSSKTFE IDLDQPTDFS ARYKERRSAQ 540
541 TAQPELKSET NEIRSKELQL TKSSSATELR NSPGLVAVSA AKQKIATETE TETAERPINY 600
601 CEEGTPGSFS RFDSLNSLTE KPEKCMPPKT PTKTAVLPVH VDGNTPQNID SALETPLMFS 660
661 RRSSMDSLVG DDETVACEDN GSVISEYSRM QSGVISPSEL PDSPTQSMPQ SPRRDRKVST 720
721 QNNLDTPEQK PSTVFEDKLN RFHVEHTPAA FSCATSLSNL SMMDDSNANA IRGQRGNDIN 780
781 GNGDAPRSYC TEDTTAVLSK APSNSDLSIL SIPNDLNANE AQPVPAPRAD VTGMDTRMPA 840
841 EDAISKMRCG GNALPSYLPV SDEMSKYYVE DSPCTFSVIS GLSHLTVGSA KAGPVLKLPM 900
901 RTAEEAQAPK LPPRRSAVQG DAEPRLPPKK SDSLSSLSMD SDDDCNLLSQ AIAAGSCRPQ 960
961 PSGASTSSSL ANASTSTLCR ENGQSKKQVE HGDKPNYSSD DSLDDDDDDA RSKSLFEQCI1020
1021 LSGMHKSNDA LESEGEPPGQ RQEISARDRF VSNQVRQIES MLAGRQH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
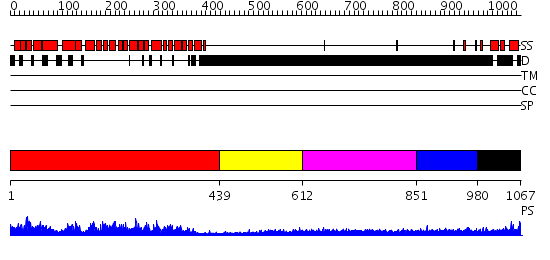
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..438] | 98.69897 | No description for 2gl7A was found. | |
| 2 | View Details | [439..611] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [612..850] | 2.69 | Beta-catenin in complex with a phosphorylated APC 20aa repeat fragment | |
| 4 | View Details | [851..979] | 4.197995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [980..1067] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)