













| General Information: |
|
| Name(s) found: |
Dcr-1-PA /
FBpp0083717
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2249 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] RNA-induced silencing complex [IDA] |
| Biological Process: |
pre-microRNA processing
[IDA][IMP][TAS]
gene silencing by miRNA [TAS] production of miRNAs involved in gene silencing by miRNA [IMP][TAS] siRNA loading onto RISC involved in RNA interference [IMP] germ-line stem cell division [IMP] pole cell formation [IMP] segment polarity determination [IGI] production of siRNA involved in RNA interference [IDA] RNA interference [IDA][IMP][TAS] dsRNA transport [IMP] mitotic cell cycle, embryonic [IMP] dendrite morphogenesis [IMP] |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IPI] double-stranded RNA binding [IEA][ISS] helicase activity [IEA] bidentate ribonuclease III activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAFHWCDNNL HTTVFTPRDF QVELLATAYE RNTIICLGHR SSKEFIALKL LQELSRRARR 60
61 HGRVSVYLSC EVGTSTEPCS IYTMLTHLTD LRVWQEQPDM QIPFDHCWTD YHVSILRPEG 120
121 FLYLLETREL LLSSVELIVL EDCHDSAVYQ RIRPLFENHI MPAPPADRPR ILGLAGPLHS 180
181 AGCELQQLSA MLATLEQSVL CQIETASDIV TVLRYCSRPH EYIVQCAPFE MDELSLVLAD 240
241 VLNTHKSFLL DHRYDPYEIY GTDQFMDELK DIPDPKVDPL NVINSLLVVL HEMGPWCTQR 300
301 AAHHFYQCNE KLKVKTPHER HYLLYCLVST ALIQLYSLCE HAFHRHLGSG SDSRQTIERY 360
361 SSPKVRRLLQ TLRCFKPEEV HTQADGLRRM RHQVDQADFN RLSHTLESKC RMVDQMDQPP 420
421 TETRALVATL EQILHTTEDR QTNRSAARVT PTPTPAHAKP KPSSGANTAQ PRTRRRVYTR 480
481 RHHRDHNDGS DTLCALIYCN QNHTARVLFE LLAEISRRDP DLKFLRCQYT TDRVADPTTE 540
541 PKEAELEHRR QEEVLKRFRM HDCNVLIGTS VLEEGIDVPK CNLVVRWDPP TTYRSYVQCK 600
601 GRARAAPAYH VILVAPSYKS PTVGSVQLTD RSHRYICATG DTTEADSDSD DSAMPNSSGS 660
661 DPYTFGTARG TVKILNPEVF SKQPPTACDI KLQEIQDELP AAAQLDTSNS SDEAVSMSNT 720
721 SPSESSTEQK SRRFQCELSS LTEPEDTSDT TAEIDTAHSL ASTTKDLVHQ MAQYREIEQM 780
781 LLSKCANTEP PEQEQSEAER FSACLAAYRP KPHLLTGASV DLGSAIALVN KYCARLPSDT 840
841 FTKLTALWRC TRNERAGVTL FQYTLRLPIN SPLKHDIVGL PMPTQTLARR LAALQACVEL 900
901 HRIGELDDQL QPIGKEGFRA LEPDWECFEL EPEDEQIVQL SDEPRPGTTK RRQYYYKRIA 960
961 SEFCDCRPVA GAPCYLYFIQ LTLQCPIPEE QNTRGRKIYP PEDAQQGFGI LTTKRIPKLS1020
1021 AFSIFTRSGE VKVSLELAKE RVILTSEQIV CINGFLNYTF TNVLRLQKFL MLFDPDSTEN1080
1081 CVFIVPTVKA PAGGKHIDWQ FLELIQANGN TMPRAVPDEE RQAQPFDPQR FQDAVVMPWY1140
1141 RNQDQPQYFY VAEICPHLSP LSCFPGDNYR TFKHYYLVKY GLTIQNTSQP LLDVDHTSAR1200
1201 LNFLTPRYVN RKGVALPTSS EETKRAKREN LEQKQILVPE LCTVHPFPAS LWRTAVCLPC1260
1261 ILYRINGLLL ADDIRKQVSA DLGLGRQQIE DEDFEWPMLD FGWSLSEVLK KSRESKQKES1320
1321 LKDDTINGKD LADVEKKPTS EETQLDKDSK DDKVEKSAIE LIIEGEEKLQ EADDFIEIGT1380
1381 WSNDMADDIA SFNQEDDDED DAFHLPVLPA NVKFCDQQTR YGSPTFWDVS NGESGFKGPK1440
1441 SSQNKQGGKG KAKGPAKPTF NYYDSDNSLG SSYDDDDNAG PLNYMHHNYS SDDDDVADDI1500
1501 DAGRIAFTSK NEAETIETAQ EVEKRQKQLS IIQATNANER QYQQTKNLLI GFNFKHEDQK1560
1561 EPATIRYEES IAKLKTEIES GGMLVPHDQQ LVLKRSDAAE AQVAKVSMME LLKQLLPYVN1620
1621 EDVLAKKLGD RRELLLSDLV ELNADWVARH EQETYNVMGC GDSFDNYNDH HRLNLDEKQL1680
1681 KLQYERIEIE PPTSTKAITS AILPAGFSFD RQPDLVGHPG PSPSIILQAL TMSNANDGIN1740
1741 LERLETIGDS FLKYAITTYL YITYENVHEG KLSHLRSKQV ANLNLYRLGR RKRLGEYMIA1800
1801 TKFEPHDNWL PPCYYVPKEL EKALIEAKIP THHWKLADLL DIKNLSSVQI CEMVREKADA1860
1861 LGLEQNGGAQ NGQLDDSNDS CNDFSCFIPY NLVSQHSIPD KSIADCVEAL IGAYLIECGP1920
1921 RGALLFMAWL GVRVLPITRQ LDGGNQEQRI PGSTKPNAEN VVTVYGAWPT PRSPLLHFAP1980
1981 NATEELDQLL SGFEEFEESL GYKFRDRSYL LQAMTHASYT PNRLTDCYQR LEFLGDAVLD2040
2041 YLITRHLYED PRQHSPGALT DLRSALVNNT IFASLAVRHG FHKFFRHLSP GLNDVIDRFV2100
2101 RIQQENGHCI SEEYYLLSEE ECDDAEDVEV PKALGDVFES IAGAIFLDSN MSLDVVWHVY2160
2161 SNMMSPEIEQ FSNSVPKSPI RELLELEPET AKFGKPEKLA DGRRVRVTVD VFCKGTFRGI2220
2221 GRNYRIAKCT AAKCALRQLK KQGLIAKKD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
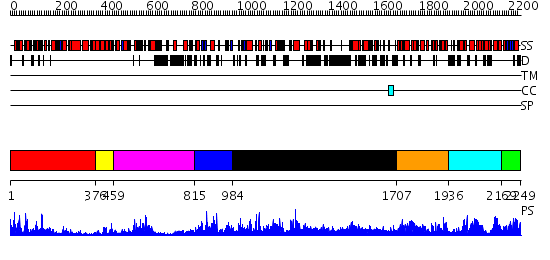
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..375] | 69.39794 | No description for 2j0qA was found. | |
| 2 | View Details | [376..458] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [459..814] | 14.69897 | Nucleotide excision repair enzyme UvrB | |
| 4 | View Details | [815..983] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [984..1706] | 2.1 | Crystal Structure of Dicer from Giardia intestinalis | |
| 6 | View Details | [1707..1935] | 57.69897 | RNase III endonuclease catalytic domain; RNase III, C-terminal domain | |
| 7 | View Details | [1936..2168] | 31.39794 | RNase III endonuclease catalytic domain | |
| 8 | View Details | [2169..2249] | 6.045757 | No description for 2yt4A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)