













| General Information: |
|
| Name(s) found: |
Dcr-2-PA /
FBpp0086061
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1722 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
siRNA loading onto RISC involved in RNA interference
[IMP]
defense response to virus [IMP] chromatin silencing [IMP] production of siRNA involved in RNA interference [IDA][IMP][TAS] RNA interference [IDA][IMP][NAS][TAS] dsRNA transport [IMP] targeting of mRNA for destruction involved in RNA interference [IMP] |
| Molecular Function: |
ATP binding
[IEA]
ribonuclease III activity [IEA] double-stranded RNA binding [IEA][ISS][NAS] ATP-dependent helicase activity [IEA] helicase activity [ISS][NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDVEIKPRG YQLRLVDHLT KSNGIVYLPT GSGKTFVAIL VLKRFSQDFD KPIESGGKRA 60
61 LFMCNTVELA RQQAMAVRRC TNFKVGFYVG EQGVDDWTRG MWSDEIKKNQ VLVGTAQVFL 120
121 DMVTQTYVAL SSLSVVIIDE CHHGTGHHPF REFMRLFTIA NQTKLPRVVG LTGVLIKGNE 180
181 ITNVATKLKE LEITYRGNII TVSDTKEMEN VMLYATKPTE VMVSFPHQEQ VLTVTRLISA 240
241 EIEKFYVSLD LMNIGVQPIR RSKSLQCLRD PSKKSFVKQL FNDFLYQMKE YGIYAASIAI 300
301 ISLIVEFDIK RRQAETLSVK LMHRTALTLC EKIRHLLVQK LQDMTYDDDD DNVNTEEVIM 360
361 NFSTPKVQRF LMSLKVSFAD KDPKDICCLV FVERRYTCKC IYGLLLNYIQ STPELRNVLT 420
421 PQFMVGRNNI SPDFESVLER KWQKSAIQQF RDGNANLMIC SSVLEEGIDV QACNHVFILD 480
481 PVKTFNMYVQ SKGRARTTEA KFVLFTADKE REKTIQQIYQ YRKAHNDIAE YLKDRVLEKT 540
541 EPELYEIKGH FQDDIDPFTN ENGAVLLPNN ALAILHRYCQ TIPTDAFGFV IPWFHVLQED 600
601 ERDRIFGVSA KGKHVISINM PVNCMLRDTI YSDPMDNVKT AKISAAFKAC KVLYSLGELN 660
661 ERFVPKTLKE RVASIADVHF EHWNKYGDSV TATVNKADKS KDRTYKTECP LEFYDALPRV 720
721 GEICYAYEIF LEPQFESCEY TEHMYLNLQT PRNYAILLRN KLPRLAEMPL FSNQGKLHVR 780
781 VANAPLEVII QNSEQLELLH QFHGMVFRDI LKIWHPFFVL DRRSKENSYL VVPLILGAGE 840
841 QKCFDWELMT NFRRLPQSHG SNVQQREQQP APRPEDFEGK IVTQWYANYD KPMLVTKVHR 900
901 ELTPLSYMEK NQQDKTYYEF TMSKYGNRIG DVVHKDKFMI EVRDLTEQLT FYVHNRGKFN 960
961 AKSKAKMKVI LIPELCFNFN FPGDLWLKLI FLPSILNRMY FLLHAEALRK RFNTYLNLHL1020
1021 LPFNGTDYMP RPLEIDYSLK RNVDPLGNVI PTEDIEEPKS LLEPMPTKSI EASVANLEIT1080
1081 EFENPWQKYM EPVDLSRNLL STYPVELDYY YHFSVGNVCE MNEMDFEDKE YWAKNQFHMP1140
1141 TGNIYGNRTP AKTNANVPAL MPSKPTVRGK VKPLLILQKT VSKEHITPAE QGEFLAAITA1200
1201 SSAADVFDME RLEILGDSFL KLSATLYLAS KYSDWNEGTL TEVKSKLVSN RNLLFCLIDA1260
1261 DIPKTLNTIQ FTPRYTWLPP GISLPHNVLA LWRENPEFAK IIGPHNLRDL ALGDEESLVK1320
1321 GNCSDINYNR FVEGCRANGQ SFYAGADFSS EVNFCVGLVT IPNKVIADTL EALLGVIVKN1380
1381 YGLQHAFKML EYFKICRADI DKPLTQLLNL ELGGKKMRAN VNTTEIDGFL INHYYLEKNL1440
1441 GYTFKDRRYL LQALTHPSYP TNRITGSYQE LEFIGDAILD FLISAYIFEN NTKMNPGALT1500
1501 DLRSALVNNT TLACICVRHR LHFFILAENA KLSEIISKFV NFQESQGHRV TNYVRILLEE1560
1561 ADVQPTPLDL DDELDMTELP HANKCISQEA EKGVPPKGEF NMSTNVDVPK ALGDVLEALI1620
1621 AAVYLDCRDL QRTWEVIFNL FEPELQEFTR KVPINHIRQL VEHKHAKPVF SSPIVEGETV1680
1681 MVSCQFTCME KTIKVYGFGS NKDQAKLSAA KHALQQLSKC DA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
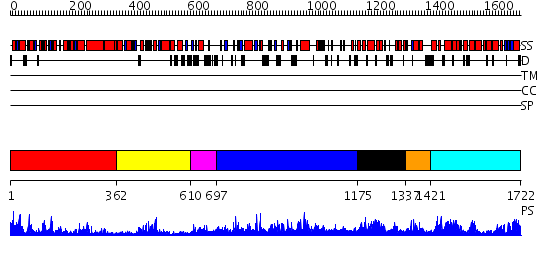
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..361] | 85.0 | No description for 2i4iA was found. | |
| 2 | View Details | [362..609] | 19.69897 | Initiation factor 4a | |
| 3 | View Details | [610..696] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [697..1174] | 3.88 | Crystal structure of full length Argonaute from Pyrococcus furiosus | |
| 5 | View Details | [1175..1336] | 12.0 | Crystal Structure of Dicer from Giardia intestinalis | |
| 6 | View Details | [1337..1420] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1421..1722] | 44.045757 | RNase III endonuclease catalytic domain; RNase III, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)