













| General Information: |
|
| Name(s) found: |
BCL10_MOUSE
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Mus musculus |
| Length: | 233 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[UNKNOWN][IDA]
membrane raft [IDA] membrane [IEA] cytosol [UNKNOWN][IDA] protein complex [UNKNOWN] intracellular [IEA] immunological synapse [IDA] |
| Biological Process: |
protein homooligomerization
[IMP]
[NOT]activation of JUN kinase activity [UNKNOWN] regulation of apoptosis [IEA] protein heterooligomerization [UNKNOWN] response to fungus [IMP] mast cell activation [NAS] I-kappaB kinase/NF-kappaB cascade [UNKNOWN][IMP] positive regulation of NF-kappaB transcription factor activity [UNKNOWN][IMP] cellular defense response [IMP] positive regulation of T cell activation [IMP] neural tube closure [IMP] induction of apoptosis [UNKNOWN] [NOT]apoptosis [UNKNOWN][IDA][IMP] immunoglobulin mediated immune response [IMP] regulation of T cell receptor signaling pathway [IMP] activation of caspase activity [UNKNOWN] |
| Molecular Function: |
protein homodimerization activity
[UNKNOWN]
protein binding [IPI] protein heterodimerization activity [UNKNOWN] caspase activator activity [UNKNOWN] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEAPAPSLTE EDLTEVKKDA LENLRVYLCE KIIAERHFDH LRAKKILSRE DTEEISCRTS 60
61 SRKRAGKLLD YLQENPRGLD TLVESIRREK TQSFLIQKIT DEVLKLRNIK LEHLKGLKCS 120
121 SCEPFAAGAT NNLSRCNSDE SNLSEKQRAS TVMYHPEGES STAPFFSMAS SLNLPVLEVG 180
181 RTENSSFSSA TLPRPGDPGA PPLPPDLRLE EGGSCGNSSE MFLPLRSRAL SRQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
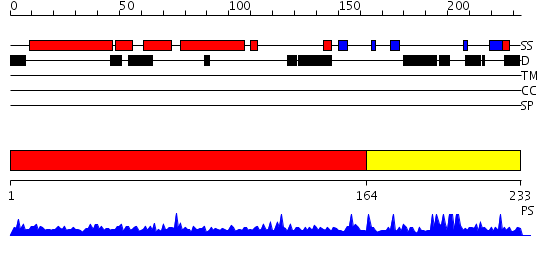
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..163] | 2.68 | NMR STRUCTURE OF THE RAIDD CARD DOMAIN, 15 STRUCTURES | |
| 2 | View Details | [164..233] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)