













| General Information: |
|
| Name(s) found: |
gi|5689463
[NCBI NR]
gi|20140914 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 593 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 AAAARGRCLP CSLPSAIAQP HLCPASALCS RRPGLGQPGQ PPSSPHAALA SSSPPSPPPP 60
61 ARGRALPPMV SRPEPEGEAM DAELAVAPPG CSHLGSFKVD NWKQNLRAIY QCFVWSGTAE 120
121 ARKRKAKSCI CHVCGVHLNR LHSCLYCVFF GCFTKKHIHE HAKAKRHNLA IDLMYGGIYC 180
181 FLCQDYIYDK DMEIIAKEEQ RKAWKMQGVG EKFSTWEPTK RELELLKHNP KRRKITSNCT 240
241 IGLRGLINLG NTCFMNCIVQ ALTHTPLLRD FFLSDRHRCE MQSPSSCLVC EMSSLFQEFY 300
301 SGHRSPHIPY KLLHLVWTHA RHLAGYEQQD AHEFLIAALD VLHRHCKGDD NGKKANNPNH 360
361 CNCIIDQIFT GGLQSDVTCQ VCHGVSTTID PFWDISLDLP GSSTPFWPLS PGSEGNVVNG 420
421 ESHVSGTTTL TDCLRRFTRP EHLGSSAKIK CSGCHSYQES TKQLTMKKLP IVACFHLKRF 480
481 EHSAKLRRKI TTYVSFPLEL DMTPFMASSK ESRMNGQYQQ PTDSLNNDNK YSLFAVVNHQ 540
541 GTLESGHYTS FIRQHKDQWF KCDDAIITKA SIKDVLDSEG YLLFYHKQFL EYE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
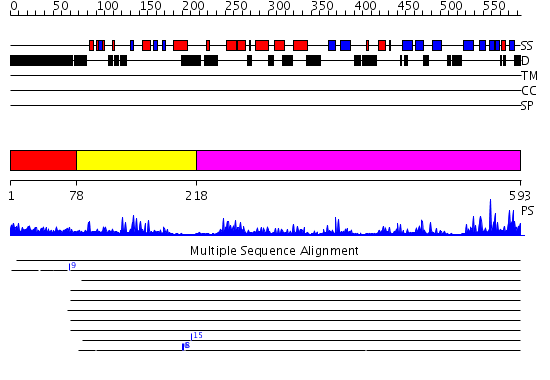
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [78..217] | 11.69897 | No description for 2g43A was found. | |
| 3 | View Details | [218..593] | 62.045757 | No description for 2ibiA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.90 |
Source: Reynolds et al. (2008)