













| General Information: |
|
| Name(s) found: |
PNKP
[HGNC (HUGO)]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Homo sapiens |
| Length: | 521 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGEVEAPGRL WLESPPGGAP PIFLPSDGQA LVLGRGPLTQ VTDRKCSRTQ VELVADPETR 60
61 TVAVKQLGVN PSTTGTQELK PGLEGSLGVG DTLYLVNGLH PLTLRWEETR TPESQPDTPP 120
121 GTPLVSQDEK RDAELPKKRM RKSNPGWENL EKLLVFTAAG VKPQGKVAGF DLDGTLITTR 180
181 SGKVFPTGPS DWRILYPEIP RKLRELEAEG YKLVIFTNQM SIGRGKLPAE EFKAKVEAVV 240
241 EKLGVPFQVL VATHAGLYRK PVTGMWDHLQ EQANDGTPIS IGDSIFVGDA AGRPANWAPG 300
301 RKKKDFSCAD RLFALNLGLP FATPEEFFLK WPAAGFELPA FDPRTVSRSG PLCLPESRAL 360
361 LSASPEVVVA VGFPGAGKST FLKKHLVSAG YVHVNRDTLG SWQRCVTTCE TALKQGKRVA 420
421 IDNTNPDAAS RARYVQCARA AGVPCRCFLF TATLEQARHN NRFREMTDSS HIPVSDMVMY 480
481 GYRKQFEAPT LAEGFSAILE IPFRLWVEPR LGRLYCQFSE G |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
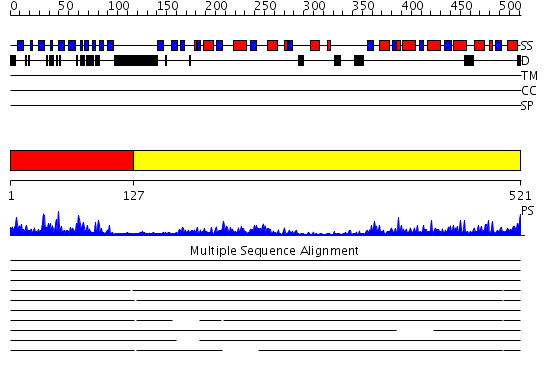
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..126] | 29.09691 | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme | |
| 2 | View Details | [127..521] | 72.30103 | Molecular architecture of mammalian polynucleotide kinase, a DNA repair enzyme |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)