













| General Information: |
|
| Name(s) found: |
HOS1 /
YPR068C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 470 amino acids |
Gene Ontology: |
|
| Cellular Component: |
histone deacetylase complex
[TAS |
| Biological Process: |
regulation of transcription, DNA-dependent
[ISS protein amino acid deacetylation [ISS chromatin organization [ISS |
| Molecular Function: |
histone deacetylase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKLVISTSI FQSQVADLLP CNNHQKSQLT YSLINAYDLL QHFDEVLTFP YARKDDLLEF 60
61 HSKSYIDYLI NGRFNKMMAQ DVNNPMVESK WSELSELADN WNEKIDYNPS QDLQRFTTRE 120
121 NLYNYYLNHS QALENNMDCI NNSEVPTNDK PTDTYILNSE TKQYNLEGDC PIFSYLPMYC 180
181 QVITGATLNL LDHLSPTERL IGINWDGGRH HAFKQRASGF CYINDVVLLI QRLRKAKLNK 240
241 ITYVDFDLHH GDGVEKAFQY SKQIQTISVH LYEPGFFPGT GSLSDSRKDK NVVNIPLKHG 300
301 CDDNYLELIA SKIVNPLIER HEPEALIIEC GGDGLLGDRF NEWQLTIRGL SRIIINIMKS 360
361 YPRAHIFLLG GGGYNDLLMS RFYTYLTWCV TKQFSNLRCG DNNSFQIDPF DVCDGDDSEQ 420
421 FIREHDLVEM YNEENYQYWI YEMEGSSRMK MLRNDNKDRD MVELMKFYEL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
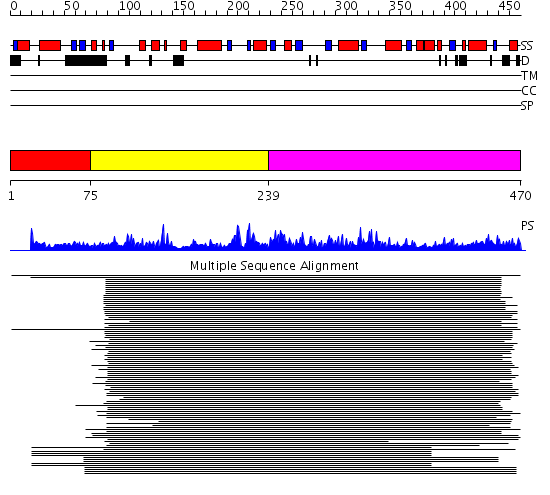
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [75..238] | 280.228787 | HDAC homologue | |
| 3 | View Details | [239..470] | 280.228787 | HDAC homologue |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)