













| General Information: |
|
| Name(s) found: |
GLO4 /
YOR040W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 285 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA mitochondrion [IDA |
| Biological Process: |
carbohydrate metabolic process
[IMP methylglyoxal catabolic process to D-lactate [IDA telomere maintenance [IMP |
| Molecular Function: |
hydroxyacylglutathione hydrolase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKFLLQQIRN MHVKPIKMRW LTGGVNYSYL LSTEDRRNSW LIDPAEPLEV SPKLSAEEKK 60
61 SIDAIVNTHH HYDHSGGNLA LYSILCQENS GHDIKIIGGS KSSPGVTEVP DNLQQYHLGN 120
121 LRVTCIRTPC HTKDSICYYI KDLETGEQCI FTGDTLFIAG CGRFFEGTGR DMDMALNQIM 180
181 LRAVGETNWN KVKIYPGHEY TKGNVSFIRA KIYSDIGQNK EFDALEQYCK SNECTTGHFT 240
241 LRDELGYNPF MRLDDRAVRL AVGDTAGTYP RSVVMQELRK LKNAM |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
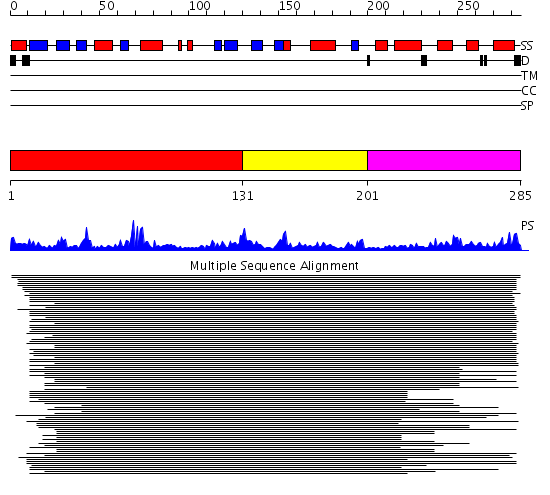
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..130] | 393.29073 | Glyoxalase II (hydroxyacylglutathione hydrolase) | |
| 2 | View Details | [131..200] | 393.29073 | Glyoxalase II (hydroxyacylglutathione hydrolase) | |
| 3 | View Details | [201..285] | 393.29073 | Glyoxalase II (hydroxyacylglutathione hydrolase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)