













| General Information: |
|
| Name(s) found: |
YNL040W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 456 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPTPMTPVKV GALACQRNSF LFDGFKTLVV SCEPTKNKKG EIEGYEIELQ DTILFPEGGG 60
61 QPSDSGFLKI VEGNRNSSKI EKILVSHVSR FGLHAKHHVN DYIEPGTTVE VAVDEQKRMD 120
121 YMQQHTGQHL LSAILERNYK VDTVSWSMGG IITKKKPVLE PSDYFNYIEL NRKLTLDEIT 180
181 NVSDEINQLI INFPQEIIVE ERIGEETVDE VSTSKIPDDY DLSKGVLRTI HIGDIDSNPC 240
241 CGTHLKCTSQ IGSILILSNQ SAVRGSNSRL YFMCGKRVSL YAKSVNKILL DSKNLLSCSE 300
301 TQISEKITRQ TKQIQQLNKR EQYWIKRLAR TASEELMNTL KASGKKRAYF MEEEYGTLEL 360
361 LLQIHKEVSN FLKDDTEGYE IILCGYERQT NTGSLLILSE SGEKIANLAA NLGSILQNLK 420
421 GGGGKKGGKW QGKITSISNA EFAALSDYLS HDFASC |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #15 Mitotic 2nd-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
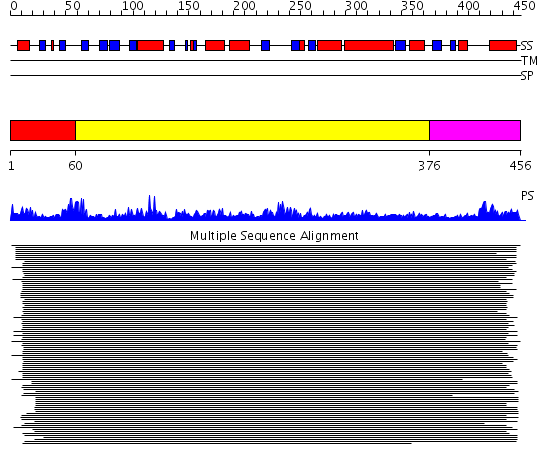
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..59] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [60..375] | 50.69897 | Threonyl-tRNA synthetase (ThrRS), C-terminal domain; Threonyl-tRNA synthetase (ThrRS), N-terminal 'additional' domain; Threonyl-tRNA synthetase (ThrRS), second 'additional' domain; Threonyl-tRNA synthetase (ThrRS) | |
| 3 | View Details | [376..456] | 4.097997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)