













| General Information: |
|
| Name(s) found: |
NDI1 /
YML120C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 513 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA mitochondrion [IDA |
| Biological Process: |
mitochondrial electron transport, NADH to ubiquinone
[IDA positive regulation of apoptosis [IMP NADH oxidation [IDA chronological cell aging [IMP |
| Molecular Function: |
NADH dehydrogenase (ubiquinone) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSKNLYSNK RLLTSTNTLV RFASTRSTGV ENSGAGPTSF KTMKVIDPQH SDKPNVLILG 60
61 SGWGAISFLK HIDTKKYNVS IISPRSYFLF TPLLPSAPVG TVDEKSIIEP IVNFALKKKG 120
121 NVTYYEAEAT SINPDRNTVT IKSLSAVSQL YQPENHLGLH QAEPAEIKYD YLISAVGAEP 180
181 NTFGIPGVTD YGHFLKEIPN SLEIRRTFAA NLEKANLLPK GDPERRRLLS IVVVGGGPTG 240
241 VEAAGELQDY VHQDLRKFLP ALAEEVQIHL VEALPIVLNM FEKKLSSYAQ SHLENTSIKV 300
301 HLRTAVAKVE EKQLLAKTKH EDGKITEETI PYGTLIWATG NKARPVITDL FKKIPEQNSS 360
361 KRGLAVNDFL QVKGSNNIFA IGDNAFAGLP PTAQVAHQEA EYLAKNFDKM AQIPNFQKNL 420
421 SSRKDKIDLL FEENNFKPFK YNDLGALAYL GSERAIATIR SGKRTFYTGG GLMTFYLWRI 480
481 LYLSMILSAR SRLKVFFDWI KLAFFKRDFF KGL |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | experimental2 - mih1-2 | McCusker D, et al (2007) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
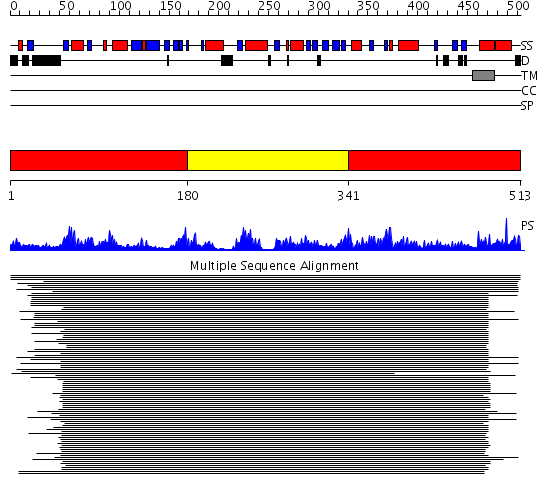
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..179] [341..513] |
101.0 | Dihydrolipoamide dehydrogenase | |
| 2 | View Details | [180..340] | 101.0 | Dihydrolipoamide dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|