













| General Information: |
|
| Name(s) found: |
CIR2 /
YOR356W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 631 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
NAD catabolic process
[IPI |
| Molecular Function: |
oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIKFTNENLI RGIRMTISAK SRHLALGTDM TRKFSLSCRF LNKANLTEEE KELLNEPRAR 60
61 DYVDVCIVGG GPAGLATAIK LKQLDNSSGT GQLRVVVLEK SSVLGGQTVS GAILEPGVWK 120
121 ELFPDEKSDI GIPLPKELAT LVTKEHLKFL KGKWAISVPE PSQMINKGRN YIVSLNQVVG 180
181 YLGEKAEEVG VEVYPGIAVS DLIYDENNAV KGVITKDAGI SKSGKPKETF ERGMEFWARQ 240
241 TVLAEGCHGS LTKQALAKYD LRKGRQHQTY GLGIKEVWEV KPENFNKGFA AHTMGYPLTN 300
301 DVYGGGFQYH FGDGLVTVGL VVGLDYKNPY VSPYKEFQKM KHHPYYSKVL EGGKCIAYAA 360
361 RALNEGGLQS VPKLNFPGGV LVGASAGFMN VPKIKGTHTA MKSGLLAAES IFESIKGLPV 420
421 LEEVEDEDAK MAMFDKEATI NLESYESAFK ESSIYKELYE VRNIRPSFSG KLGGYGGMIY 480
481 SGIDSLILKG KVPWTLKFDE KNDGEILEPA SKYKPIEYPK PDGVISFDIL TSVSRTGTYH 540
541 DDDEPCHLRV PGQDMVKYAE RSFPVWKGVE SRFCPAGVYE FVKDEKSPVG TRLQINSQNC 600
601 IHCKTCDIKA PRQDITWKVP EGGDGPKYTL T |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
AIM45 ALD4 CIR1 CIR2 CIT1 CYB2 DLD1 FUM1 GUT2 MDH1 NDE1 NDE2 NDI1 SDH1 |
| View Details | Gavin AC, et al. (2002) |

|
CIR2 PUS7 |
| View Details | Qiu et al. (2008) |

|
CIR2 GUT2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
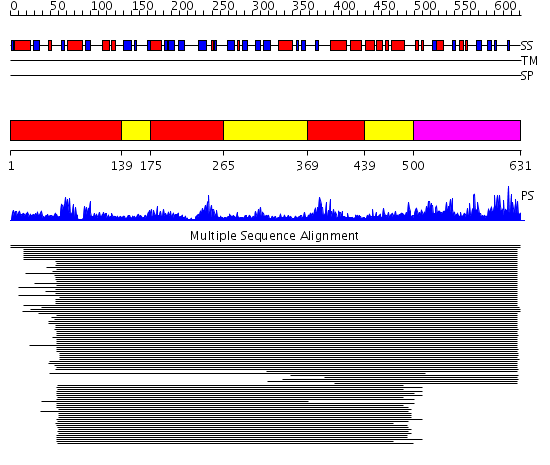
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..138] [175..264] [369..438] |
39.69897 | p-Hydroxybenzoate hydroxylase, PHBH; p-Hydroxybenzoate hydroxylase (PHBH) | |
| 2 | View Details | [139..174] [265..368] [439..499] |
39.69897 | p-Hydroxybenzoate hydroxylase, PHBH; p-Hydroxybenzoate hydroxylase (PHBH) | |
| 3 | View Details | [500..631] | 13.069997 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 | |
| 2 | No functions predicted. |
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)