













| General Information: |
|
| Name(s) found: |
DLD1 /
YDL174C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 587 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial inner membrane [IDA |
| Biological Process: |
carbohydrate metabolic process
[IMP aerobic respiration [IMP |
| Molecular Function: |
D-lactate dehydrogenase (cytochrome) activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLWKRTCTRL IKPIAQPRGR LVRRSCYRYA STGTGSTDSS SQWLKYSVIA SSATLFGYLF 60
61 AKNLYSRETK EDLIEKLEMV KKIDPVNSTL KLSSLDSPDY LHDPVKIDKV VEDLKQVLGN 120
121 KPENYSDAKS DLDAHSDTYF NTHHPSPEQR PRIILFPHTT EEVSKILKIC HDNNMPVVPF 180
181 SGGTSLEGHF LPTRIGDTIT VDLSKFMNNV VKFDKLDLDI TVQAGLPWED LNDYLSDHGL 240
241 MFGCDPGPGA QIGGCIANSC SGTNAYRYGT MKENIINMTI VLPDGTIVKT KKRPRKSSAG 300
301 YNLNGLFVGS EGTLGIVTEA TVKCHVKPKA ETVAVVSFDT IKDAAACASN LTQSGIHLNA 360
361 MELLDENMMK LINASESTDR CDWVEKPTMF FKIGGRSPNI VNALVDEVKA VAQLNHCNSF 420
421 QFAKDDDEKL ELWEARKVAL WSVLDADKSK DKSAKIWTTD VAVPVSQFDK VIHETKKDMQ 480
481 ASKLINAIVG HAGDGNFHAF IVYRTPEEHE TCSQLVDRMV KRALNAEGTC TGEHGVGIGK 540
541 REYLLEELGE APVDLMRKIK LAIDPKRIMN PDKIFKTDPN EPANDYR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
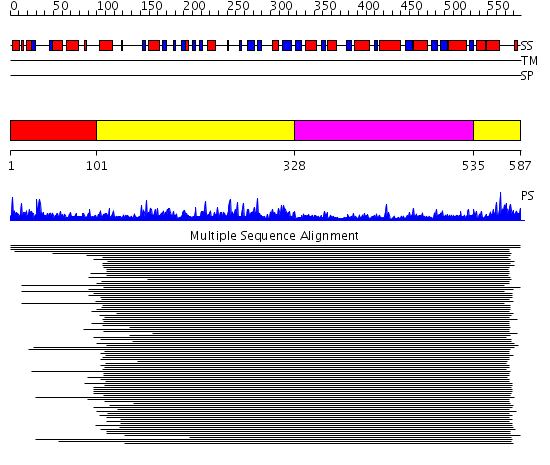
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [101..327] [535..587] |
105.69897 | Flavoprotein subunit of p-cresol methylhydroxylase | |
| 3 | View Details | [328..534] | 105.69897 | Flavoprotein subunit of p-cresol methylhydroxylase |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)