













| General Information: |
|
| Name(s) found: |
DPH2 /
YKL191W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 534 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
peptidyl-diphthamide biosynthetic process from peptidyl-histidine
[IMP ribosome biogenesis [RCA |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEVAPALSTT QSDVAFQKVE THEIDRSSYL GPCYNSDELM QLISAYYNVE PLVGYLEQHP 60
61 EYQNVTLQFP DDLIKDSSLI VRLLQSKFPH GKIKFWVLAD TAYSACCVDE VAAEHVHAEV 120
121 VVHFGDACLN AIQNLPVVYS FGTPFLDLAL VVENFQRAFP DLSSKICLMA NAPFSKHLSQ 180
181 LYNILKGDLH YTNIIYSQVN TSAVEEKFVT ILDTFHVPED VDQVGVFEKN SVLFGQHDKA 240
241 DNISPEDYHL FHLTTPQDPR LLYLSTVFQS VHIFDPALPG MVTGPFPSLM RRYKYMHVAR 300
301 TAGCIGILVN TLSLRNTRET INELVKLIKT REKKHYLFVV GKPNVAKLAN FEDIDIWCIL 360
361 GCSQSGIIVD QFNEFYKPII TPYELNLALS EEVTWTGKWV VDFRDAIDEI EQNLGGQDTI 420
421 SASTTSDEPE FDVVRGRYTS TSRPLRALTH LELEAADDDD SKQLTTRHTA SGAVIKGTVS 480
481 TSASALQNRS WKGLGSDFDS TEVDNTGADI EEGISGVARG YGFDREDAMK KENK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ATS1 DPH1 DPH2 EFT2, EFT1 KTI11 |
| View Details | Krogan NJ, et al. (2006) |
|
AIM22 DPH1 DPH2 EFT2, EFT1 HGH1 MNN1 PGI1 RRG10 TRX1 YMR315W |
| View Details | Gavin AC, et al. (2006) |
|
DPH1 DPH2 EFT2, EFT1 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
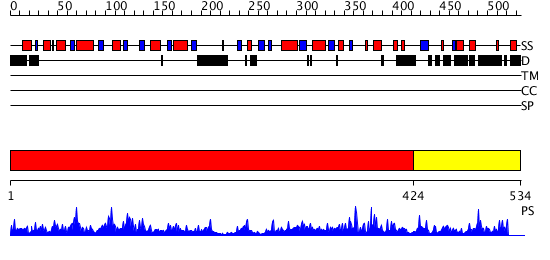
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..423] | 13.39794 | Putative diphthamide synthesis protein No confident structure predictions are available. | |
| 2 | View Details | [424..534] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)