













| General Information: |
|
| Name(s) found: |
IBA57 /
YJR122W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 497 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFISRRCRIK GFTLKNLLWF RSSSTRFVST ESPDASAITK PDGIFNYSLL ENRTYIRIRG 60
61 PDTVKFLNGL VTSKLLPHFI KKNLTTVEEN EVPTEEGTTK VDPIIPVPEF DARLGNWGLY 120
121 NEKGIQGPYI SRFGLYSAFL NGKGKLITDT IIYPTPVTVS EQISNYPEYL LELHGNVVDK 180
181 ILHVLQTHKL ANKIKFEKID HSSLKTWDVE VQFPNLPKDI ENPWFDNLLD PMALPKNSID 240
241 ANNFAVNVLN SLFNSDPRIL GIYVERRTES MSRHYSTFPQ SFRVVTSEQV DDLSKLFNFN 300
301 VFDFPFQVNK KASVQVREIR FQKGLIDSTE DYISETLLPL ELNFDFFPNT ISTNKGCYVG 360
361 QELTARTYAT GILRKRLVPV KLDNYQLLDT DPERKYAEFH IDNVVEKSLA ENEPTLNPFT 420
421 NKPPERTKRK QRPAGLLISN EGLYGVALLR TEHFSAAFSS DEPVEFYITT TKGENIKITP 480
481 QKPFWFSDWK NNNGPHK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
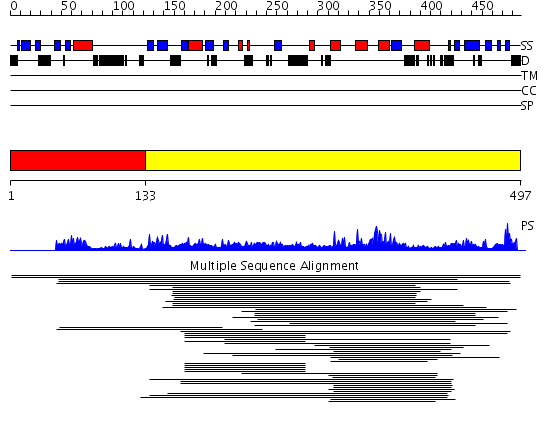
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [133..497] | 12.047997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [432..497] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)