













| General Information: |
|
| Name(s) found: |
GLG2 /
YJL137C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 380 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
glycogen biosynthetic process
[IGI |
| Molecular Function: |
glycogenin glucosyltransferase activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKKVAICTL LYSRDYLPGA LTLAYQLQKL LKHAVVEDEI TLCLLIEKKL FGDEFKPQEI 60
61 ALIRSLFKEI IIIEPLKDQE KSIEKNKANL ELLKRPELSH TLLKARLWEL VQFDQVLFLD 120
121 ADTLPLNKEF FEILRLYPEQ TRFQIAAVPD IGWPDMFNTG VLLLIPDLDM ATSLQDFLIK 180
181 TVSIDGADQG IFNQFFNPIC NYSKEVLHKV SPLMEWIRLP FTYNVTMPNY GYQSSPAMNF 240
241 FQQHIRLIHF IGTFKPWSRN TTDYDDHYYQ LWRSTQRELY SECHLSNYFT HLQLGNIETE 300
301 TNFYHEPPCL QDLLNHGKRE NQKHVDLDIT SVDRNASQKS TAEKHDIEKP TSKPQSAFKF 360
361 DWESTDYLDR VQRAFPKPDT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
ADE5,7 DAP1 GLG1 GLG2 GSY1 GSY2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #19 Asynchronous Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #19b Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) | |
| View Run | #19a Asynchronous Prep1-new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
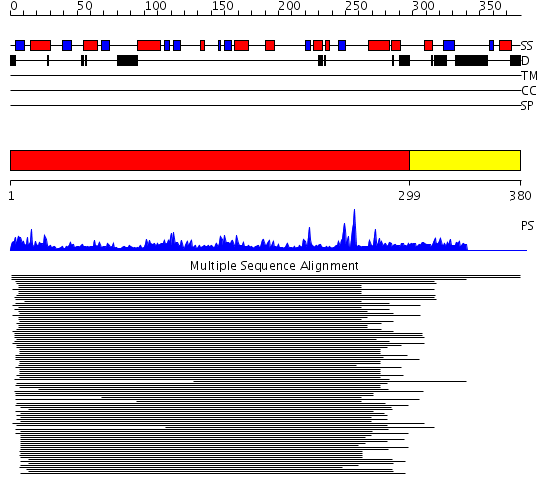
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..298] | 279.218487 | Glycogenin | |
| 2 | View Details | [299..380] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)