













| General Information: |
|
| Name(s) found: |
KSP1 /
YHR082C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1029 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
protein amino acid phosphorylation
[IDA |
| Molecular Function: |
protein serine/threonine kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLDYEIYKE GGILNNRYQK IEDISEGSYG YVSLAKDVRE KRLVAVKYIF KLEDDGQYDG 60
61 PQDDENDCDS SDCDDDEDTK VDTDRHENEN GNASSNNGSS REKKHNLYKH KKSLISSKVK 120
121 SRLSNNICLE AMYEVDIQTK IGRHQNIAAL LDFFDSYIIM EYCSGGDLYE AIKADAVPKK 180
181 TKSITHIITQ IMDAIEYVHN KGIYHRDIKP ENILISGIDW TIKLTDWGLA TTDKTSMDRN 240
241 VGSERYMSPE LFDSNLDIKE RKEPYDCAKV DLWAMGIVFL NIVFHKNPFS IANQSDKSFC 300
301 YFAANREALF DVFSTMAYDF FQVLRYSLTI DPANRDLKMM RTELQNLSEY TLDDEYYNNL 360
361 DEGYEETMID GLPPQPVPPS SAPVSLPTPI SSSNKQHMPE FKKDFNFNNV NERKRSDVSQ 420
421 NQNVASGFFK KPSTQQQKFF NQGYNTTLST HERAKSAPKF KFKKRNKYGR TDNQFSKPVN 480
481 IEDRKKSKIL KKSRKPLGIP TPNTHMNNFF HDYKARDEFN TRDFFTPPSV QHRYMEGFSN 540
541 NNNKQYRQNR NYNNNNNNSN NNHGSNYNNF NNGNSYIKGW NKNFNKYRRP SSSSYTGKSP 600
601 LSRYNMSYNH NNNSSINGYA RRGSTTTVQH SPGAYIPPNA RNHHVSPTNQ FLRVPQSTAP 660
661 DISTVLGGKP SYQEHYTQDS MDSEGDHDSD DVLFTLEEGD HDFVNGMDNL SINDHLPHTT 720
721 VGSHNEVFVH ASTNHNNNGN NNHIDTNSTT NQYHRQYIPP PLTTSLHINN NNNESNELPD 780
781 LLKSPASSEA HLNLSSGPID PILTGNIGNR YSHSSDSKEL EQERRLSMEQ KFKNGVYVPP 840
841 HHRKSFNLGT QVPPMNMKTS NEATLSVSHN SVNFGGSYNS RRSSANESNP LHMNKALEKL 900
901 SSSPGAKSSF VGFPKPLLPR NHSSTTIALQ NEDVFADSNN DAIIFEDEEY EGESDKMAHG 960
961 KMEGGDNESS STSPDERQIF GPYEIYAQTF AGSTHDKKLG AGRKTSIQDE MVGSLEQYKN1020
1021 NWLILQQQD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
KSP1 RIA1 SDO1 |
| View Details | Ho Y, et al. (2002) |
|
ARO1 BCK1 CHS1 CMP2 DBP7 KOG1 KSP1 PRI2 PSY2 TPD3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
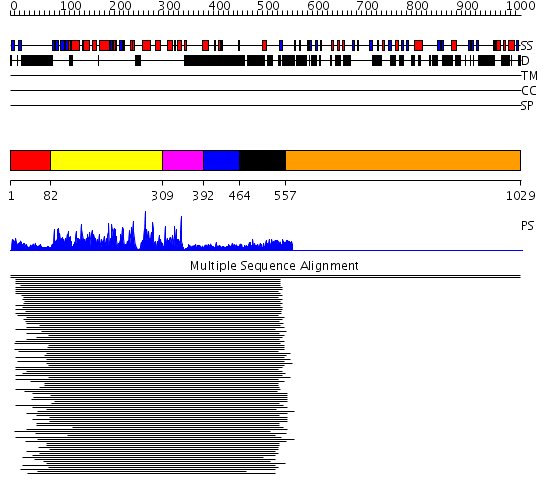
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..81] | 13.0 | Chk2 kinase | |
| 2 | View Details | [82..308] | 95.228787 | Type I TGF-beta receptor R4 | |
| 3 | View Details | [309..391] | 273.9897 | Calmodulin-dependent protein kinase | |
| 4 | View Details | [392..463] | 16.30103 | Calmodulin | |
| 5 | View Details | [464..556] | 16.30103 | Calmodulin | |
| 6 | View Details | [557..1029] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)