













| General Information: |
|
| Name(s) found: |
PHB1 /
YGR132C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 287 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial inner membrane [IDA |
| Biological Process: |
negative regulation of proteolysis
[IMP protein folding [IDA replicative cell aging [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNSAKLIDV ITKVALPIGI IASGIQYSMY DVKGGSRGVI FDRINGVKQQ VVGEGTHFLV 60
61 PWLQKAIIYD VRTKPKSIAT NTGTKDLQMV SLTLRVLHRP EVLQLPAIYQ NLGLDYDERV 120
121 LPSIGNEVLK SIVAQFDAAE LITQREIISQ KIRKELSTRA NEFGIKLEDV SITHMTFGPE 180
181 FTKAVEQKQI AQQDAERAKF LVEKAEQERQ ASVIRAEGEA ESAEFISKAL AKVGDGLLLI 240
241 RRLEASKDIA QTLANSSNVV YLPSQHSGGG NSESSGSPNS LLLNIGR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #16 Mitotic-New search criteria | Keck JM, et al. (2011) | |
| View Run | #18 Mitotic Prep4-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
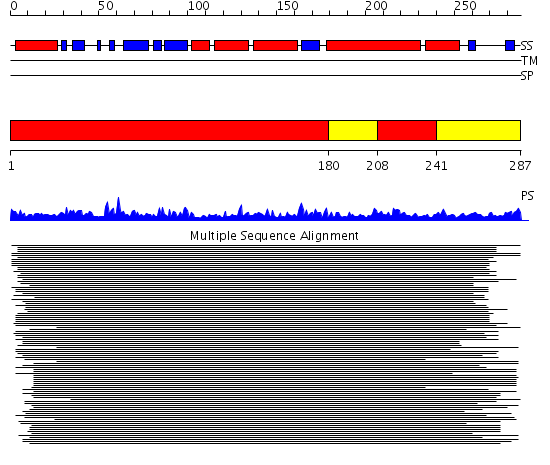
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..179] [208..240] |
877.529817 | No description for 1lu7A_ was found. | |
| 2 | View Details | [180..207] [241..287] |
877.529817 | No description for 1lu7A_ was found. | |
| 3 | View Details | [198..287] | 3.672997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 |
|
||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)