













| General Information: |
|
| Name(s) found: |
RMD8 /
YFR048W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 662 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
response to drug
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYKANQPSP GEMPKRSPSI LVTDARTSKN RMSAPFAGHA AGSRKNMENA GVTKSQGVRS 60
61 SAIGPSPLQS FTHPRRRSSG RFSDISIDNI LSDNSDIPSA RREERLSSSS SDRPRQYERL 120
121 SSRRKMINPL PPRTSKTSQK LVLIPEDDNL NHFQTLPTNA LDRQRPKVGS MKSNSFDRLP 180
181 RYSKEKSMAR ITAYNVADGF NLNQLYKFLQ ETHEVSPRLY DECLYVAYTL PLLPGKGGFR 240
241 IKSNLSKKTM GGKTLIDNLI DTSEQRDHHY EYYSGVETVE DANNNYELET SGNNNNANQD 300
301 TTTVPDHLPN PVGQQDSFNP MEPQFFAEET PLEIEKRERT ERINMLKKEE NDSDASCGND 360
361 NNNKNNDKSK LYAVEGNDQY VQSSRSPASP SSISTPSPPS SSQNDFDRVY KMHRDNDHEG 420
421 NDRHAEIFIF HYGVIVFWNF TEIQEKNILG DITFADYKNL MIRPLDEQDI ETEQFHFEYD 480
481 RDTERPRIFN DIVTLRSGDH IIELTLSHAI AQSSKLSRFE SRISPILISV TKLPKRLALY 540
541 GTLGLKREQL LKKSGKLFKL RVDVNLSSTI LDTPEFFWSF EPSLHPLYVA MREYLEIDQR 600
601 VQVLNDRCKV FLEFFDICVD SVAERNMARV TWWFILVILF GVIFSLTEIF VRYVIIHRHT 660
661 ST |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
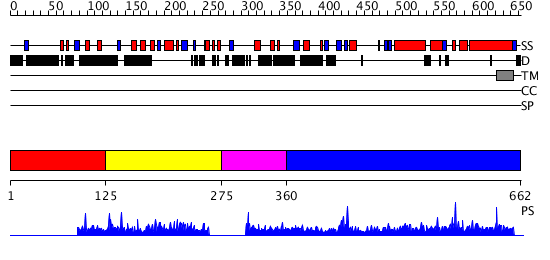
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..124] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [125..274] | 1.002966 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [275..359] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [360..662] | 148.091515 | Uncharacterised ACR, YagE family COG1723 No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|