













| General Information: |
|
| Name(s) found: |
ROG3 /
YFR022W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 733 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGFSSGKSTK KKPLLFDIRL KNVDNDVILL KGPPNEAPSV LLSGCIVLSI NEPMQIKSIS 60
61 LRLYGKIQID VPLERPQDAS SSSLSSSPPK IRKYNKVFYN YAWDNVNLKE YLSGLRGQSG 120
121 LAGSSSSSNI LGTRQRAQST SSLKSLKGSS SPSSCTLDKG NYDFPFSAIL PGSLPESVES 180
181 LPNCFVTYSM ESVIERSKNY SDLICRKNIR VLRTISPAAV ELSETVCVDN SWPDKVDYSI 240
241 SVPNKAVAIG SATPINISIV PLSKGLKLGS IKVVLFENYQ YCDPFPPVIS ENRQVTELNL 300
301 EDPLNESSGE FNGNGCFVNN PFFQPDHSFQ DKWEIDTILQ IPNSLSNCVQ DCDVRSNIKV 360
361 RHKLKFFIIL INPDGHKSEL RASLPIQLFI SPFVALSIKP LSSSNLYSLF STTNQKDENS 420
421 SQEEEEEYLF SRSASVTGLE LLADMRSGGS VPTISDLMTP PNYEMHVYDR LYSGSFTRTA 480
481 VETSGTCTPL GSECSTVEDQ QQDLEDLRIR LTKIRNQRDN LGLPPSASSA AASRSLSPLL 540
541 NVPAPEDGTE RILPQSALGP NSGSVPGVHS NVSPVLLSRS PAPSVSAHEV LPVPSGLNYP 600
601 ETQNLNKVPS YGKAMKYDII GEDLPPSYPC AIQNVQPRKP SRVHSRNSST TLSSSIPTSF 660
661 HSSSFMSSTA SPISIINGSR SSSSGVSLNT LNELTSKTSN NPSSNSMKRS PTRRRATSLA 720
721 GFMGGFLSKG NKR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
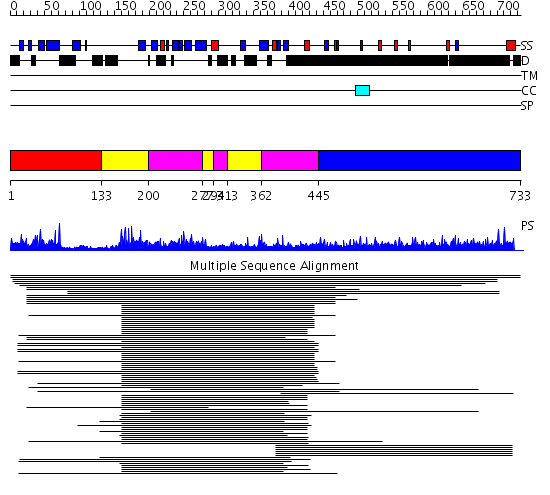
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..132] | 2.065995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [133..199] [277..293] [313..361] |
64.522879 | Arrestin | |
| 3 | View Details | [200..276] [294..312] [362..444] |
64.522879 | Arrestin | |
| 4 | View Details | [445..733] | 20.136997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)