













| General Information: |
|
| Name(s) found: |
CBS2 /
YDR197W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 389 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial inner membrane [TAS |
| Biological Process: |
translation
[TAS |
| Molecular Function: |
molecular_function
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSIPRVYS LGNSAMTYLL ALRIAQLPSQ PKVPSVVLLL NDQKKLNRFL NNDSKIIVKS 60
61 SNNNKETYHR QFMASCVPPI LSNGEIAPIE NLIVSDPSSK FITAQLSKYN KSLRPETNIL 120
121 FLNPSLNLLE HLHRYRWRFD EARPNLFMGF TTPVDVGTIH QEFQLSLKVK GRIQFHIAKI 180
181 DGFPRMSSTG KSASLSLRGD RQKNEKENNA FYKLFREISR LRSGIGSDLV SFDLHVHGFQ 240
241 DLFFTELEKL ILESCTEPLL AVYDCVYKKE LLKIPGAQDI IKKLISEQLS IIDRSYPSLN 300
301 TNPNYSVIFD KERIFSLVMR DLEVNGHKRA KLAQSLNQLN QTNINELNGF FVSLGKYKKC 360
361 NCKWNDILLT LIKGKQFITK QKALDYHYL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
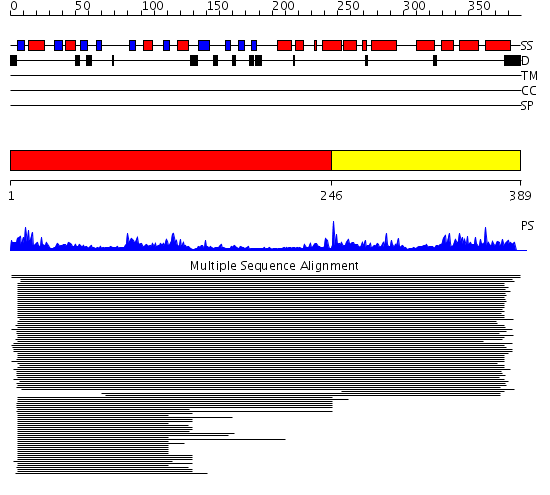
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..245] | 57.522879 | Ketopantoate reductase PanE | |
| 2 | View Details | [246..389] | 57.522879 | Ketopantoate reductase PanE |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.59 |
Source: Reynolds et al. (2008)