













| General Information: |
|
| Name(s) found: |
SCC2 /
YDR180W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1493 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear cohesin complex
[TAS |
| Biological Process: |
mitotic sister chromatid cohesion
[IMP double-strand break repair via homologous recombination [IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSYPGKDKNI PGRIIEALED LPLSYLVPKD GLAALVNAPM RVSLPFDKTI FTSADDGRDV 60
61 NINVLGTANS TTSSIKNEAE KERLVFKRPS NFTSSANSVD YVPTNFLEGL SPLAQSVLST 120
121 HKGLNDSINI EKKSEIVSRP EAKHKLESVT SNAGNLSFND NSSNKKTKTS TGVTMTQANL 180
181 AEQYLNDLKN ILDIVGFDQN SAEIGNIEYW LQLPNKKFVL TTNCLTKLQM TIKNITDNPQ 240
241 LSNSIEITWL LRLLDVMVCN IKFSKSSLKM GLDDSMLRYI ALLSTIVLFN IFLLGKNDSN 300
301 LHRESYIMEP VNFLSDLIES LKILTIEYGS LKIEFDTFQE ALELLPKYIR NGPFLDDNVT 360
361 AKLVYIFSDL LMNNDIEATT NIQFQSFWDN VKRISSDILV SLFGSFDQQR GFIIEELLSH 420
421 IEKLPTKRIQ KKLRKVGNQN IYITDFTFTL MSMLENINCY SFCNQMKDIA PENIDLLKNE 480
481 YKKQEEFLFN IVEHINDTIL ERFFKNPSAL RYVIDNFVQD LLLLISSPQW PVTEKILSSL 540
541 LKRLLSVYSP SMQVSANIET ICLQLIGNIG STIFDIKCST RDHEDNNLIK MINYPETLPH 600
601 FFKSFEECIA YNETIKCRRS ATRFLWNLRL GTILILEEYT KDAKEQIITV DNELKKILEQ 660
661 IKDGGLGPEL ENREADFSTI KLDYFSILHA FELLNLYDPY LKLILSLLAK DKIKLRSTAI 720
721 KCLSMLASKD KVILSNPMVK ETIHRRLNDS SASVKDAILD LVSINSSYFE FYQQINNNYN 780
781 DDSIMVRKHV LRINEKMYDE TNDIVTKVYV IARILMKIED EEDNIIDMAR LILLNRWILK 840
841 VHEVLDQPEK LKEISSSVLL VMSRVAIMNE KCSQLFDLFL NFYLLNKEAH SKEAYDKITH 900
901 VLTILTDFLV QKIVELNSDD TNEKNSIVDK QNFLNLLAKF ADSTVSFLTK DHITALYPYM 960
961 VSDEKSDFHY YILQVFRCTF EKLANFKQKF LYDLETTLLS RLPKMNVREI DEAMPLIWSV1020
1021 ATHRHDTARV AKACSSCLSH LHPYINKANN EEAAIVVDGK LQRLIYLSTG FARFCFPKPS1080
1081 NDKIAFLQEG ETLYEHITKC LLVLSKDKIT HVIRRVAVKN LTKLCGNHPK LFNSRHVLHL1140
1141 LDKEFQSDQL DIKLVILESL YDLFLLEERK SVRNTGVNST LSSNSILKKK LLKTNRVEFA1200
1201 NDGVCSALAT RFLDNILQLC LLRDLKNSLV AIRLLKLILK FGYTNPSHSI PTVIALFAST1260
1261 SQYIRHVAYE LLEDLFEKYE TLVFSSLSRG VTKAIHYSIH TDEKYYYKHD HFLSLLEKLC1320
1321 GTGKKNGPKF FKVLKRIMQS YLDDITDLTS TNSSVQKSIF VLCTNISNIT FVSQYDLVSL1380
1381 LKTIDLTTDR LKEVIMDEIG DNVSSLSVSE EKLSGIILIQ LSLQDLGTYL LHLYGLRDDV1440
1441 LLLDIVEESE LKNKQLPAKK PDISKFSAQL ENIEQYSSNG KLLTYFRKHV KDT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
IRR1 MCD1 REC8 SCC2 SMC1 SMC3 |
| View Details | Krogan NJ, et al. (2006) |
|
MDJ1 SCC2 SCC4 |
| View Details | Qiu et al. (2008) |
|
IRR1 MCD1 SCC2 SMC1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3908: TAP-tagged, includes hir2(delta) in background | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
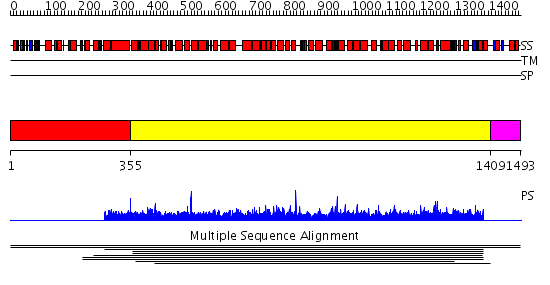
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..354] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [355..1408] | 1.007903 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [1409..1493] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [381..480] | 1.033993 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [481..1309] | 3.13 | No description for 2ie3A was found. | |
| 6 | View Details | [1310..1493] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)