













| General Information: |
|
| Name(s) found: |
HIS4 /
YCL030C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 799 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[TAS |
| Biological Process: |
histidine biosynthetic process
[TAS |
| Molecular Function: |
phosphoribosyl-ATP diphosphatase activity
[TAS phosphoribosyl-AMP cyclohydrolase activity [TAS histidinol dehydrogenase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLPILPLID DLASWNSKKE YVSLVGQVLL DGSSLSNEEI LQFSKEEEVP LVALSLPSGK 60
61 FSDDEIIAFL NNGVSSLFIA SQDAKTAEHL VEQLNVPKER VVVEENGVFS NQFMVKQKFS 120
121 QDKIVSIKKL SKDMLTKEVL GEVRTDRPDG LYTTLVVDQY ERCLGLVYSS KKSIAKAIDL 180
181 GRGVYYSRSR NEIWIKGETS GNGQKLLQIS TDCDSDALKF IVEQENVGFC HLETMSCFGE 240
241 FKHGLVGLES LLKQRLQDAP EESYTRRLFN DSALLDAKIK EEAEELTEAK GKKELSWEAA 300
301 DLFYFALAKL VANDVSLKDV ENNLNMKHLK VTRRKGDAKP KFVGQPKAEE EKLTGPIHLD 360
361 VVKASDKVGV QKALSRPIQK TSEIMHLVNP IIENVRDKGN SALLEYTEKF DGVKLSNPVL 420
421 NAPFPEEYFE GLTEEMKEAL DLSIENVRKF HAAQLPTETL EVETQPGVLC SRFPRPIEKV 480
481 GLYIPGGTAI LPSTALMLGV PAQVAQCKEI VFASPPRKSD GKVSPEVVYV AEKVGASKIV 540
541 LAGGAQAVAA MAYGTETIPK VDKILGPGNQ FVTAAKMYVQ NDTQALCSID MPAGPSEVLV 600
601 IADEDADVDF VASDLLSQAE HGIDSQVILV GVNLSEKKIQ EIQDAVHNQA LQLPRVDIVR 660
661 KCIAHSTIVL CDGYEEALEM SNQYAPEHLI LQIANANDYV KLVDNAGSVF VGAYTPESCG 720
721 DYSSGTNHTL PTYGYARQYS GANTATFQKF ITAQNITPEG LENIGRAVMC VAKKEGLDGH 780
781 RNAVKIRMSK LGLIPKDFQ |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
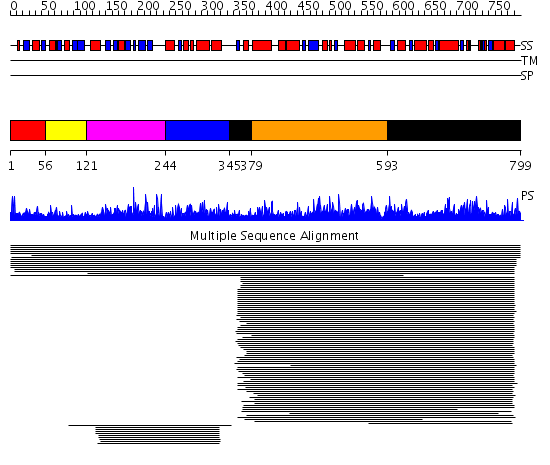
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..55] | 3.009987 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [56..120] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [121..243] | 50.124939 | Phosphoribosyl-AMP cyclohydrolase No confident structure predictions are available. | |
| 4 | View Details | [244..344] | 39.744727 | Phosphoribosyl-ATP pyrophosphohydrolase No confident structure predictions are available. | |
| 5 | View Details | [345..378] [593..799] |
1145.9897 | L-histidinol dehydrogenase HisD | |
| 6 | View Details | [379..592] | 1145.9897 | L-histidinol dehydrogenase HisD |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 |
|
||||||||||||||||||
| 4 |
|
||||||||||||||||||
| 5 |
|
||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)