













| General Information: |
|
| Name(s) found: |
HSL7 /
YBR133C
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 827 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular bud neck
[IDA |
| Biological Process: |
G2/M transition of mitotic cell cycle
[IGI |
| Molecular Function: |
protein-arginine N-methyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHSNVFVGVK PGFNHKQHSK KSRFLENVSS HSPELPSNYD YVLLPITTPR YKEIVGQVFK 60
61 DFQRQSIQNW KPLQIPEPQL QDICIPPFNV KKLDNDDTPS YIGLLSSWLE LESRDPNVRD 120
121 LGLKVLLNEC KYARFVGINK LILAPPRDLS NLQLYGQMIY RLLQNRIVFA APALTISISL 180
181 PLYEDSDPLA TWELWNTVRK QCEYHPSLTI SLALPRTRTP SYVLNRWLAE PVSCLLVSSS 240
241 IFASNQYDYP VLHKFNQNLI LKFQKVNGDS QILGNELCVI LHGMEKYANN VKGGESAYLE 300
301 YINYLLKKGD KVLNSNSNHQ FLLQEDSRIM PPLKPHSDNL LNSTYLTFEK DLVKYDLYES 360
361 AILEALQDLA PRASAKRPLV ILVAGAGRGP LVDRTFKIIS MLFMDSKVSI IAIEKNPQAY 420
421 LYLQKRNFDC WDNRVKLIKE DMTKWQINEP SEKRIQIDLC ISELLGSFGC NELSPECLWS 480
481 IEKYHSHNDT IFIPRSYSSY IAPISSPLFY QKLSQTNRSL EAPWIVHRVP YCILSSRVNE 540
541 VWRFEHPMAQ KDTVQDEDDF TVEFSQSSLN EFKIKHRGEI HGFIGFFSAN LYNNIFLSTL 600
601 PNDSTVRLKF SEETLMNTRR EENLIKKCDH TPNMTSWSPI IFPLKQPISF IDDSELSVLM 660
661 SRIHSDTEQK VWYEWSLESF IYLMLSNYTS AVTAASMTIP RSIVTDDTKT LAHNRHYSAT 720
721 TNQKLDNQID LDQDIENEEE QGFLSNLETG WQSVQDIHGL SETAKPDHLD SINKPMFDLK 780
781 STKALEPSNE LPRHEDLEED VPEVHVRVKT SVSTLHNVCG RAFSLPL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
AHC1 CLB2 COP1 HSL7 KEL1 PRP6 SWE1 UBP15 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #27 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps3-5 | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #14 Mitotic Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) | |
| View Run | #01 Alpha factor Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #28 Asynchronous Prep5-TiO2 Phosphopeptide Enrichment | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #05 Alpha Factor Prep2-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #07 Alpha Factor Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
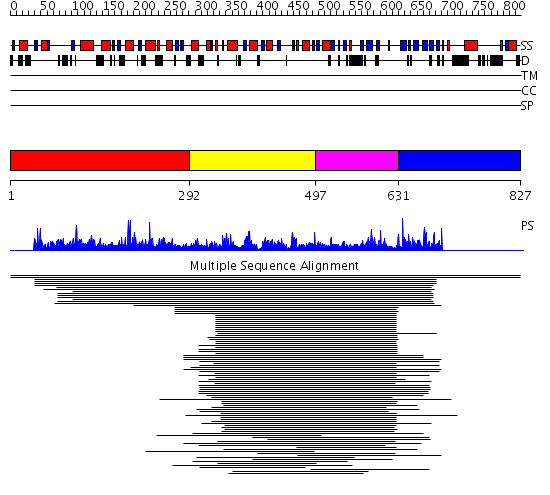
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..291] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [292..496] | 76.69897 | Arginine methyltransferase, HMT1 | |
| 3 | View Details | [497..630] | 76.69897 | Arginine methyltransferase, HMT1 | |
| 4 | View Details | [631..827] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)