













| General Information: |
|
| Name(s) found: |
DUN1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 513 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLSTKREHS GDVTDSSFKR QQRSNKPSSE YTCLGHLVNL IPGKEQKVEI TNRNVTTIGR 60
61 SRSCDVILSE PDISTFHAEF HLLQMDVDNF QRNLINVIDK SRNGTFINGN RLVKKDYILK 120
121 NGDRIVFGKS CSFLFKYASS SSTDIENDDE KVSSESRSYK NDDEVFKKPQ ISATSSQNAT 180
181 TSAAIRKLNK TRPVSFFDKY LLGKELGAGH YALVKEAKNK KTGQQVAVKI FHAQQNDDQK 240
241 KNKQFREETN ILMRVQHPNI VNLLDSFVEP ISKSQIQKYL VLEKIDDGEL FERIVRKTCL 300
301 RQDESKALFK QLLTGLKYLH EQNIIHRDIK PENILLNITR RENPSQVQLG PWDEDEIDIQ 360
361 VKIADFGLAK FTGEMQFTNT LCGTPSYVAP EVLTKKGYTS KVDLWSAGVI LYVCLCGFPP 420
421 FSDQLGPPSL KEQILQAKYA FYSPYWDKID DSVLHLISNL LVLNPDERYN IDEALNHPWF 480
481 NDIQQQSSVS LELQRLQITD NKIPKTYSEL SCL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
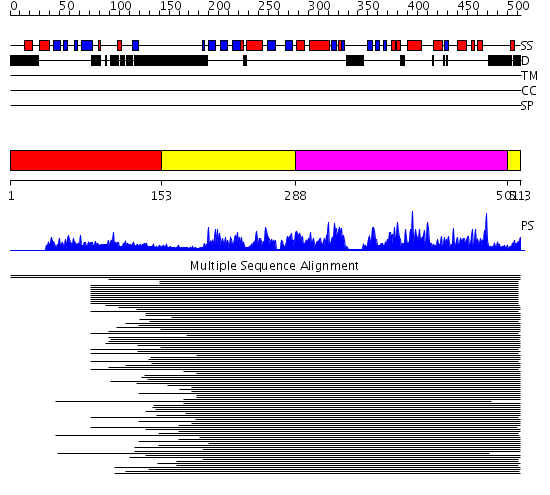
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..152] | 22.39794 | Chk2 kinase | |
| 2 | View Details | [153..287] [501..513] |
636.457575 | Calmodulin-dependent protein kinase | |
| 3 | View Details | [288..500] | 636.457575 | Calmodulin-dependent protein kinase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)