













| General Information: |
|
| Name(s) found: |
FKB53_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 477 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
nucleolus [IDA] |
| Biological Process: |
nucleosome assembly
[IMP]
|
| Molecular Function: |
peptidyl-prolyl cis-trans isomerase activity
[ISS]
FK506 binding [ISS] histone binding [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGFWGLEVKP GKPQAYNPKN EQGKIHVTQA TLGTGLSKEK SVIQCSIGDK APIALCSLLP 60
61 NKIECCPLNL EFDDDDEPVE FTVTGDRSIH LSGFLEYYQD DEDDYEHDED DSDGIDVGES 120
121 EEDDSCEYDS EEDEQLDEFE DFLDSNLERY RNAAAPKSGV IIEEIEDEEK PAKDNKAKQT 180
181 KKKSQASEGE NAKKQIVAIE GAHVPVLESE DEDEDGLPIP KGKSSEVENA SGEKMVVDND 240
241 EQGSNKKRKA KAAEQDDGQE SANKSKKKKN QKEKKKGENV LNEEAGQVQT GNVLKKQDIS 300
301 QISSNTKAQD GTANNAMSES SKTPDKSAEK KTKNKKKKKP SDEAAEISGT VEKQTPADSK 360
361 SSQVRTYPNG LIVEELSMGK PNGKRADPGK TVSVRYIGKL QKNGKIFDSN IGKSPFKFRL 420
421 GIGSVIKGWD VGVNGMRVGD KRKLTIPPSM GYGVKGAGGQ IPPNSWLTFD VELINVQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
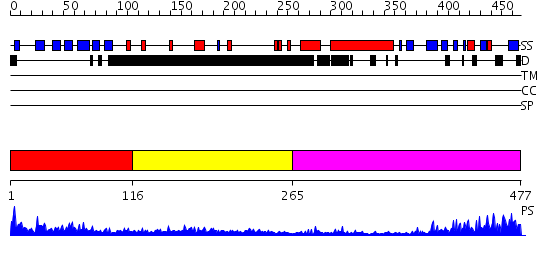
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | 2.3 | No description for 2p1bA was found. | |
| 2 | View Details | [116..264] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [265..477] | 46.69897 | Macrophage infectivity potentiator protein (MIP) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)