













| General Information: |
|
| Name(s) found: |
SSE1 /
YPL106C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 693 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
telomere maintenance
[IMP protein folding [IMP protein refolding [IDA |
| Molecular Function: |
ATP binding
[IDA chaperone regulator activity [IGI adenyl-nucleotide exchange factor activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTPFGLDLG NNNSVLAVAR NRGIDIVVNE VSNRSTPSVV GFGPKNRYLG ETGKNKQTSN 60
61 IKNTVANLKR IIGLDYHHPD FEQESKHFTS KLVELDDKKT GAEVRFAGEK HVFSATQLAA 120
121 MFIDKVKDTV KQDTKANITD VCIAVPPWYT EEQRYNIADA ARIAGLNPVR IVNDVTAAGV 180
181 SYGIFKTDLP EGEEKPRIVA FVDIGHSSYT CSIMAFKKGQ LKVLGTACDK HFGGRDFDLA 240
241 ITEHFADEFK TKYKIDIREN PKAYNRILTA AEKLKKVLSA NTNAPFSVES VMNDVDVSSQ 300
301 LSREELEELV KPLLERVTEP VTKALAQAKL SAEEVDFVEI IGGTTRIPTL KQSISEAFGK 360
361 PLSTTLNQDE AIAKGAAFIC AIHSPTLRVR PFKFEDIHPY SVSYSWDKQV EDEDHMEVFP 420
421 AGSSFPSTKL ITLNRTGDFS MAASYTDITQ LPPNTPEQIA NWEITGVQLP EGQDSVPVKL 480
481 KLRCDPSGLH TIEEAYTIED IEVEEPIPLP EDAPEDAEQE FKKVTKTVKK DDLTIVAHTF 540
541 GLDAKKLNEL IEKENEMLAQ DKLVAETEDR KNTLEEYIYT LRGKLEEEYA PFASDAEKTK 600
601 LQGMLNKAEE WLYDEGFDSI KAKYIAKYEE LASLGNIIRG RYLAKEEEKK QAIRSKQEAS 660
661 QMAAMAEKLA AQRKAEAEKK EEKKDTEGDV DMD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
FOL1 SSE1 |
| View Details | Gavin AC, et al. (2006) |

|
SSE1 TDH2 YHB1 |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
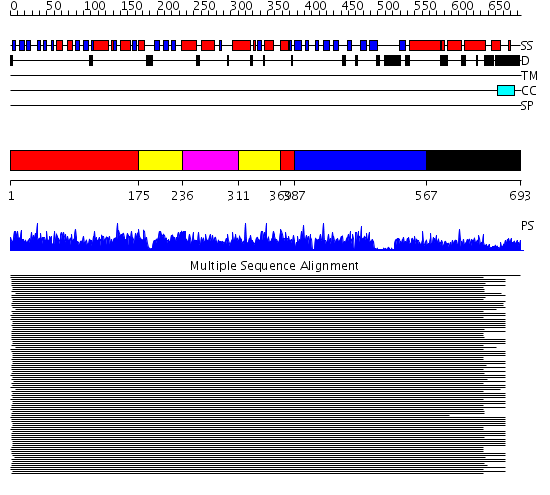
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] [369..386] |
1646.9897 | Heat shock protein 70kDa, ATPase fragment | |
| 2 | View Details | [175..235] [311..368] |
1646.9897 | Heat shock protein 70kDa, ATPase fragment | |
| 3 | View Details | [236..310] | 1646.9897 | Heat shock protein 70kDa, ATPase fragment | |
| 4 | View Details | [387..566] | 77.522879 | DnaK | |
| 5 | View Details | [567..693] | 77.522879 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)