













| General Information: |
|
| Name(s) found: |
WTM1 /
YOR230W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 437 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
regulation of meiosis
[IGI |
| Molecular Function: |
transcription corepressor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPKKVWKSST PSTYEHISSL RPKFVSRVDN VLHQRKSLTF SNVVVPDKKN NTLTSSVIYS 60
61 QGSDIYEIDF AVPLQEAASE PVKDYGDAFE GIENTSLSPK FVYQGETVSK MAYLDKTGET 120
121 TLLSMSKNGS LAWFKEGIKV PIHIVQELMG PATSYASIHS LTRPGDLPEK DFSLAISDFG 180
181 ISNDTETIVK SQSNGDEEDS ILKIIDNAGK PGEILRTVHV PGTTVTHTVR FFDNHIFASC 240
241 SDDNILRFWD TRTSDKPIWV LGEPKNGKLT SFDCSQVSNN LFVTGFSTGI IKLWDARAAE 300
301 AATTDLTYRQ NGEDPIQNEI ANFYHAGGDS VVDVQFSATS SSEFFTVGGT GNIYHWNTDY 360
361 SLSKYNPDDT IAPPQDATEE SQTKSLRFLH KGGSRRSPKQ IGRRNTAAWH PVIENLVGTV 420
421 DDDSLVSIYK PYTEESE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
RNR2 WTM1 |
| View Details | Krogan NJ, et al. (2006) |

|
GIN4 HFD1 HTZ1 KAP114 KCC4 NAP1 PRO3 REI1 RIM11 RPS6A, RPS6B UBC8 VIP1 WTM1 YKR011C |
| View Details | Gavin AC, et al. (2006) |

|
ARO3 WTM1 |
| View Details | Ho Y, et al. (2002) |
|
AAT2 ACH1 ACO1 ACS2 AFR1 AHP1 ARC1 ARF1 ATP3 CAR2 CCT2 CCT3 CCT6 CDC33 CLU1 COF1 COR1 CPR1 CYR1 CYS4 DLD3 DUG1 EGD1 ERG10 ERG13 ERG6 FPR1 FRS2 FUM1 GND1 GND2 GRX1 GUS1 HEF3 HHF1, HHF2 ILV3 LRO1 LSP1 MET6 NTF2 OLA1 OYE2 PFK1 PIL1 PRM2 RHO1 RNR2 RSN1 RVB1 SAH1 SCP160 SEC53 SEN15 SES1 SNU13 SOD1 TCP1 TEF4 THS1 TIF1, TIF2 TMA19 TSA2 UBA1 VMA4 VMA5 WTM1 WTM2 YNK1 YOR283W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #06 Alpha Factor Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
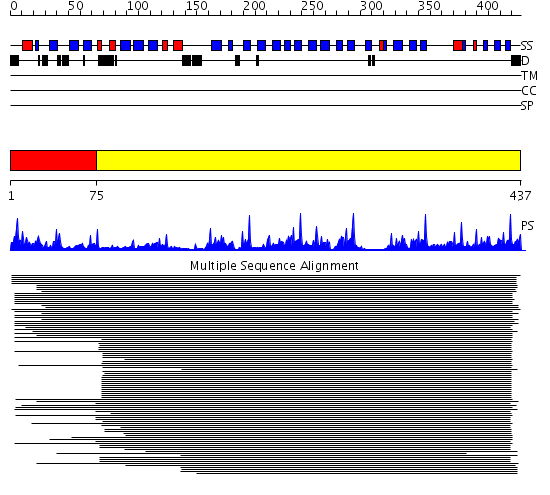
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [75..437] | 68.69897 | beta1-subunit of the signal-transducing G protein heterotrimer |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)