













| General Information: |
|
| Name(s) found: |
NSE5 /
YML023C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 556 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Smc5-Smc6 complex [IPI |
| Biological Process: |
DNA repair
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGALINSVL YVSPRNGAHY FVELTEKHLL AFEMLNSMCL LENYDHVLLF LECQFGKSHN 60
61 LAVIPFDIIL VLFTLSTLSE YYKEPILRAN DPYNTSRETL SRRALKLLQK YLAILKEFDS 120
121 EQYNLYDLEL LRCQFFLAID TLTPKKQKWG FDRFRRTKSE SGVTYRQNAS VDPELDQAKT 180
181 FKNPYRSYIS CLEQRNTILG NRLLNLKLNE PGEFINMILW TLSNSLQEST PLFLSSHEIW 240
241 MPLLEILIDL FSCRQDYFIQ HEVAQNVSKS LFVQRLSESP LAVFFESLNT RNFANRFSEY 300
301 VFLNCDYKLP SDNYATPVHP VYNGENTIVD TYIPTIKCSP LYKSQKSLAL RRKLIGSCFK 360
361 LLLRVPDGHR LITPRIVADD VIQGISRTLA SFNDILQFKK FFMTENLSQE SYFIPLLAEG 420
421 TLSEILKDTQ ECVVILTLVE NLSDGVSFCN EVIGLVKSKC FAFTEQCSQA SYEEAVLNIE 480
481 KCDVCLLVLL RYLLHLIGTE AILDAKEQLE MLHAIEKNDS GRRQWAKALN LGNDPPLLYP 540
541 IVSQMFGVHD KSVIIE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
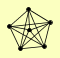
| View Details | Hazbun TR, et al. (2003) |
|
KRE29 MMS21 NSE4 NSE5 SMC5 SMC6 |
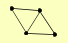
| View Details | Qiu et al. (2008) |
|
KRE29 NSE1 NSE5 SMC6 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Hazbun TR, et al. (2003) |
| View Data |
|
Unpublished Data |
| View Data |
|
Huh WK, et al. (2003) |
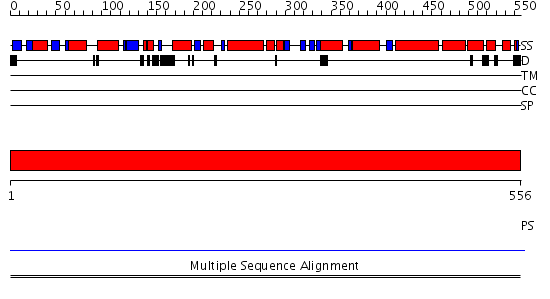
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..556] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)