













| General Information: |
|
| Name(s) found: |
SMC6 /
YLR383W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1114 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA Smc5-Smc6 complex [IPI |
| Biological Process: |
cell proliferation
[IDA DNA repair [IDA |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MISTTISGKR PIEQVDDELL SLTAQQENEE QQQQRKRRRH QFAPMTQFNS NTLDEDSGFR 60
61 SSSDVATADQ DNFLEESPSG YIKKVILRNF MCHEHFELEL GSRLNFIVGN NGSGKSAILT 120
121 AITIGLGAKA SETNRGSSLK DLIREGCYSA KIILHLDNSK YGAYQQGIFG NEIIVERIIK 180
181 RDGPASFSLR SENGKEISNK KKDIQTVVDY FSVPVSNPMC FLSQDAARSF LTASTSQDKY 240
241 SHFMKGTLLQ EITENLLYAS AIHDSAQENM ALHLENLKSL KAEYEDAKKL LRELNQTSDL 300
301 NERKMLLQAK SLWIDVAHNT DACKNLENEI SGIQQKVDEV TEKIRNRQEK IERYTSDGTT 360
361 IEAQIDAKVI YVNEKDSEHQ NARELLRDVK SRFEKEKSNQ AEAQSNIDQG RKKVDALNKT 420
421 IAHLEEELTK EMGGDKDQMR QELEQLEKAN EKLREVNNSL VVSLQDVKNE ERDIQHERES 480
481 ELRTISRSIQ NKKVELQNIA KGNDTFLMNF DRNMDRLLRT IEQRKNEFET PAIGPLGSLV 540
541 TIRKGFEKWT RSIQRAISSS LNAFVVSNPK DNRLFRDIMR SCGIRSNIPI VTYCLSQFDY 600
601 SKGRAHGNYP TIVDALEFSK PEIECLFVDL SRIERIVLIE DKNEARNFLQ RNPVNVNMAL 660
661 SLRDRRSGFQ LSGGYRLDTV TYQDKIRLKV NSSSDNGTQY LKDLIEQETK ELQNIRDRYE 720
721 EKLSEVRSRL KEIDGRLKST KNEMRKTNFR MTELKMNVGK VVDTGILNSK INERKNQEQA 780
781 IASYEAAKEE LGLKIEQIAQ EAQPIKEQYD STKLALVEAQ DELQQLKEDI NSRQSKIQKY 840
841 KDDTIYYEDK KKVYLENIKK IEVNVAALKE GIQRQIQNAC AFCSKERIEN VDLPDTQEEI 900
901 KRELDKVSRM IQKAEKSLGL SQEEVIALFE KCRNKYKEGQ KKYMEIDEAL NRLHNSLKAR 960
961 DQNYKNAEKG TCFDADMDFR ASLKVRKFSG NLSFIKDTKS LEIYILTTND EKARNVDTLS1020
1021 GGEKSFSQMA LLLATWKPMR SRIIALDEFD VFMDQVNRKI GTTLIVKKLK DIARTQTIII1080
1081 TPQDIGKIAD IDSSGVSIHR MRDPERQNNS NFYN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Hazbun TR, et al. (2003) |
|
KRE29 MMS21 NSE1 NSE3 NSE4 NSE5 SMC5 SMC6 |
| View Details | Ho Y, et al. (2002) |
|
HHF1, HHF2 IMD1 IMD4 SMC6 SRP1 |
| View Details | Qiu et al. (2008) |
|
NSE1 NSE3 NSE5 SMC5 SMC6 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #31 Asynchronous Prep (Protease cleavage) | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
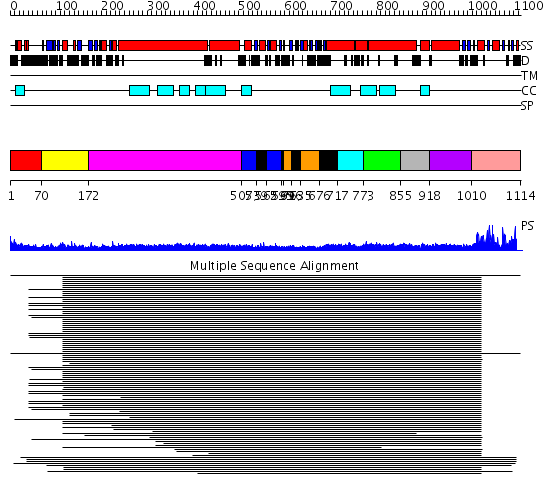
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..69] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [70..171] | 43.201249 | Rad50 | |
| 3 | View Details | [172..506] | 17.09691 | Heavy meromyosin subfragment | |
| 4 | View Details | [507..538] [562..594] |
7.56 | Smc hinge domain | |
| 5 | View Details | [539..561] [595..598] [616..634] [676..716] |
7.56 | Smc hinge domain | |
| 6 | View Details | [599..615] [635..675] |
7.56 | Smc hinge domain | |
| 7 | View Details | [717..772] | 9.522879 | Tropomyosin | |
| 8 | View Details | [773..854] | 9.522879 | Tropomyosin | |
| 9 | View Details | [855..917] | 9.522879 | Tropomyosin | |
| 10 | View Details | [918..1009] | 9.522879 | Tropomyosin | |
| 11 | View Details | [1010..1114] | 55.30103 | Smc head domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)