













| General Information: |
|
| Name(s) found: |
BYE1 /
YKL005C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 594 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter
[IGI |
| Molecular Function: |
transcription elongation regulator activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVRTSSRSN KGQNKYIEYL LQEETEAPKK KRTKKKVDSA IEKNKKSDSS QEPRKDTENV 60
61 RTDEVDEADE GYVRCLCGAN NENYDAAEYS HGDMVQCDGC DTWQHIKCMT DGKDTIDGLM 120
121 SEDSKYYCEL CDPSLYAHLE TSKEAEVSED EDYHDDVYKP VNDHDDNDAD VFLDEESPRK 180
181 RKRSPDSAKG IHIKSKQVKK SNGSKKRNKS IDAAKSDTAE NEMPTRKDFE SEKEHKLRYN 240
241 AEKMFSTLFS KFIVPETIEA KLYELPDGKD VISISQEFAH NLEEELYKAC LNVEFGTLDK 300
301 IYTEKVRSLY SNLKDKKNLE LKAHVVEGKL PLNKLVNMNA SELANPDLQE FKEKRDKIIL 360
361 ENFIVEVPDK PMYVKTHKGD ELIEDIAEPQ EDILYSKDSI RLHNIDSIDS DKSKIEQTHA 420
421 ISKEPSPSTI INEESLNCAF LYPGLGLEFT GYLNYIGASQ KLRRDIFKEA IGDGKLYVEG 480
481 RLPTTTAAPY LKEISCSRAI LVYQLFPSND SESKTTFADV VDSLENKGRI AGIKPKTRYE 540
541 KDFYIVPSKG GEIPEILKDI LGSHNDERSE RFSRMKSDER TLFAFVVVKQ EFIH |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BYE1 MLP2 NOC4 RIF1 TIF3 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
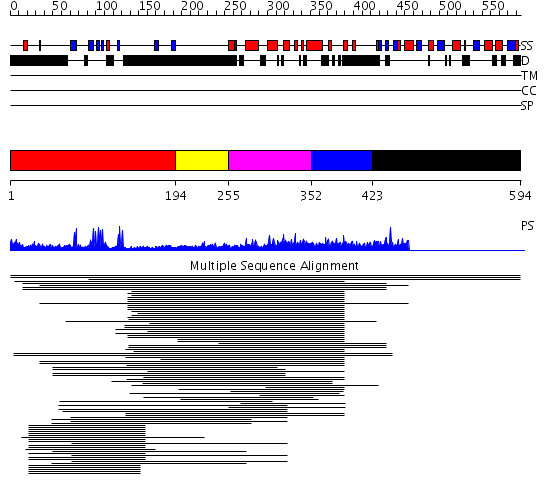
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..193] | 8.12 | Nuclear corepressor KAP-1 (TIF-1beta) | |
| 2 | View Details | [194..254] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [255..351] | 16.30103 | Elongation factor TFIIS domain 2 | |
| 4 | View Details | [352..422] | 1.049995 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [423..594] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)