













| General Information: |
|
| Name(s) found: |
egl-4 /
CE37242
[WormBase]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 470 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA cAMP-dependent protein kinase complex [IEA |
| Biological Process: |
signal transduction
[IEA protein amino acid phosphorylation [IEA |
| Molecular Function: |
ATP binding
[IEA protein tyrosine kinase activity [IEA zinc ion binding [IEA non-membrane spanning protein tyrosine kinase activity [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA cAMP-dependent protein kinase regulator activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKKVYVVSIF QNLSEDRISK MADVMDQDYY DGGHYIIRQG EKGDAFFVIN SGQVKVTQQI 60
61 EGETEPREIR VLNQGDFFGE RALLGEEVRT ANIIAQAPGV EVLTLDRESF GKLIGDLESL 120
121 KKDYGDKERL AQVVREPPSP VKIVDDFREE FAQVTLKNVK RLATLGVGGF GRVELVCVNG 180
181 DKAKTFALKA LKKKHIVDTR QQEHIFAERN IMMETSTDWI VKLYKTFRDQ KFVYMLLEVC 240
241 LGGELWTTLR DRGHFDDYTA RFYVACVLEG LEYLHRKNIV YRDLKPENCL LANTGYLKLV 300
301 DFGFAKKLAS GRKTWTFCGT PEYVSPEIIL NKGHDQAADY WALGIYICEL MLGRPPFQAS 360
361 DPMKTYTLIL KGVDALEIPN RRIGKTATAL VKKLCRDNPG ERLGSGSGGV NDIRKHRWFM 420
421 GFDWEGLRSR TLKPPILPKV SNPADVTNFD NYPPDNDVPP DEFSGWDEGF |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
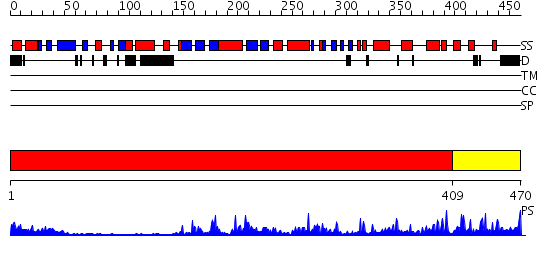
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..408] | 53.69897 | CHICKEN SRC TYROSINE KINASE | |
| 2 | View Details | [409..470] | 82.69897 | cAMP-dependent PK, catalytic subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)