













| General Information: |
|
| Name(s) found: |
HACL_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 572 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: | NONE FOUND |
| Molecular Function: |
thiamin pyrophosphate binding
[IEA]
magnesium ion binding [IEA] pyruvate decarboxylase activity [ISS] catalytic activity [IEA] transferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MADKSETTPP SIDGNVLVAK SLSHLGVTHM FGVVGIPVTS LASRAMALGI RFIAFHNEQS 60
61 AGYAASAYGY LTGKPGILLT VSGPGCVHGL AGLSNAWVNT WPMVMISGSC DQRDVGRGDF 120
121 QELDQIEAVK AFSKLSEKAK DVREIPDCVS RVLDRAVSGR PGGCYLDIPT DVLRQKISES 180
181 EADKLVDEVE RSRKEEPIRG SLRSEIESAV SLLRKAERPL IVFGKGAAYS RAEDELKKLV 240
241 EITGIPFLPT PMGKGLLPDT HEFSATAARS LAIGKCDVAL VVGARLNWLL HFGESPKWDK 300
301 DVKFILVDVS EEEIELRKPH LGIVGDAKTV IGLLNREIKD DPFCLGKSNS WVESISKKAK 360
361 ENGEKMEIQL AKDVVPFNFL TPMRIIRDAI LAVEGPSPVV VSEGANTMDV GRSVLVQKEP 420
421 RTRLDAGTWG TMGVGLGYCI AAAVASPDRL VVAVEGDSGF GFSAMEVETL VRYNLAVVII 480
481 VFNNGGVYGG DRRGPEEISG PHKEDPAPTS FVPNAGYHKL IEAFGGKGYI VETPDELKSA 540
541 LAESFAARKP AVVNVIIDPF AGAESGRLQH KN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
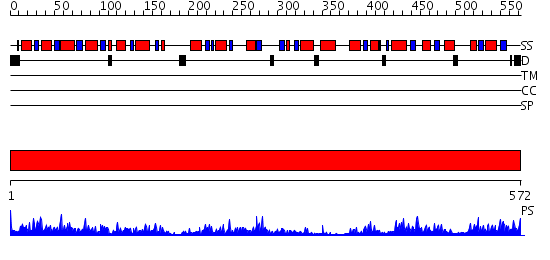
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..572] | 98.0 | CRYSTAL STRUCTURE OF OXALYL-COA DECARBOXYLASE IN COMPLEX WITH THE COFACTOR DERIVATIVE THIAMIN-2-THIAZOLONE DIPHOSPHATE AND ADENOSINE DIPHOSPHATE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|