













| General Information: |
|
| Name(s) found: |
CDKF1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 479 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] cytosol [IDA] |
| Biological Process: |
protein amino acid phosphorylation
[IDA]
maintenance of root meristem identity [IMP] regulation of cyclin-dependent protein kinase activity [TAS] |
| Molecular Function: |
cyclin-dependent protein kinase activating kinase activity
[IGI]
protein binding [IPI] protein serine/threonine kinase activity [TAS] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDKQPATSWS IHTRPEIIAK YEIFERVGSG AYADVYRARR LSDGLIVALK EIFDYQSAFR 60
61 EIDALTILNG SPNVVVMHEY FWREEENAVL VLEFLRSDLA AVIRDGKRKK KVEGGDGFSV 120
121 GEIKRWMIQI LTGVDACHRN LIVHRDLKPG NMLISDDGVL KLADFGQARI LMEHDIVASD 180
181 ENQQAYKLED KDGETSEPPE VIPDYENSPR QGSDGQEREA MSKDEYFRQV EELKAKQVVR 240
241 DDTDKDSNVH DGDISCLATC TVSEMDDDLG RNSFSYDADE AVDDTQGLMT SCVGTRWFRP 300
301 PELLYGSTMY GLEVDLWSLG CVFAELLSLE PLFPGISDID QISRVTNVLG NLNEEVWPGC 360
361 VDLPDYKSIS FAKVESPLGI EGCLPNHSGD VISLLKKLIC YDPASRATTM EMLNDKYLSE 420
421 EPLPVPVSEL YVPPTMSGPD EDSPRKWNDY REMDSDSDFD GFGPMNVKPT SSGFTIEFP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
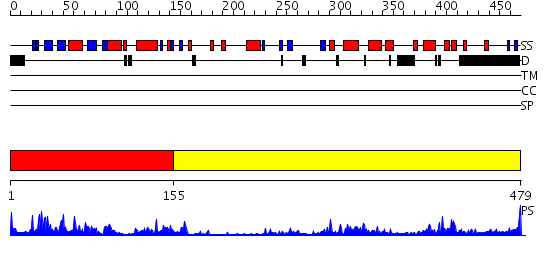
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..154] | 47.69897 | Catalytic and ubiqutin-associated domains of MARK2/PAR-1: Wild type | |
| 2 | View Details | [155..479] | 50.221849 | COORDINATES OF RAT MAP KINASE ERK2 WITH AN ARGININE MUTATION AT POSITION 52 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)