













| General Information: |
|
| Name(s) found: |
CLPC1_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 929 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast stroma
[IDA]
chloroplast [IDA] cell wall [IDA] mitochondrion [IDA] chloroplast envelope [IDA] plastid stroma [IDA] Tic complex [TAS] chloroplast thylakoid membrane [IDA] |
| Biological Process: |
regulation of chlorophyll biosynthetic process
[IMP]
protein import into chloroplast stroma [IMP] chloroplast organization [IMP] |
| Molecular Function: |
ATPase activity
[ISS]
ATP binding [ISS] ATP-dependent peptidase activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMATRVLAQ STPPSLACYQ RNVPSRGSGR SRRSVKMMCS QLQVSGLRMQ GFMGLRGNNA 60
61 LDTLGKSRQD FHSKVRQAMN VPKGKASRFT VKAMFERFTE KAIKVIMLAQ EEARRLGHNF 120
121 VGTEQILLGL IGEGTGIAAK VLKSMGINLK DARVEVEKII GRGSGFVAVE IPFTPRAKRV 180
181 LELSLEEARQ LGHNYIGSEH LLLGLLREGE GVAARVLENL GADPSNIRTQ VIRMVGENNE 240
241 VTANVGGGSS SNKMPTLEEY GTNLTKLAEE GKLDPVVGRQ PQIERVVQIL GRRTKNNPCL 300
301 IGEPGVGKTA IAEGLAQRIA SGDVPETIEG KKVITLDMGL LVAGTKYRGE FEERLKKLME 360
361 EIRQSDEIIL FIDEVHTLIG AGAAEGAIDA ANILKPALAR GELQCIGATT LDEYRKHIEK 420
421 DPALERRFQP VKVPEPTVDE TIQILKGLRE RYEIHHKLRY TDESLVAAAQ LSYQYISDRF 480
481 LPDKAIDLID EAGSRVRLRH AQVPEEAREL EKELRQITKE KNEAVRGQDF EKAGTLRDRE 540
541 IELRAEVSAI QAKGKEMSKA ESETGEEGPM VTESDIQHIV SSWTGIPVEK VSTDESDRLL 600
601 KMEETLHKRI IGQDEAVKAI SRAIRRARVG LKNPNRPIAS FIFSGPTGVG KSELAKALAA 660
661 YYFGSEEAMI RLDMSEFMER HTVSKLIGSP PGYVGYTEGG QLTEAVRRRP YTVVLFDEIE 720
721 KAHPDVFNMM LQILEDGRLT DSKGRTVDFK NTLLIMTSNV GSSVIEKGGR RIGFDLDYDE 780
781 KDSSYNRIKS LVTEELKQYF RPEFLNRLDE MIVFRQLTKL EVKEIADILL KEVFERLKKK 840
841 EIELQVTERF KERVVDEGYN PSYGARPLRR AIMRLLEDSM AEKMLAREIK EGDSVIVDVD 900
901 AEGNVTVLNG GSGTPTTSLE EQEDSLPVA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
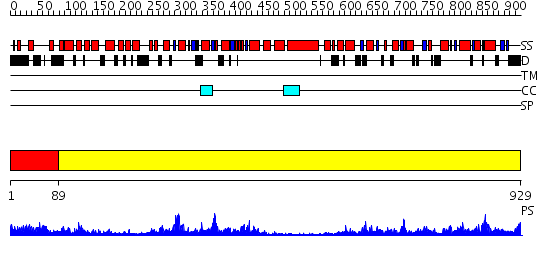
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | 80.69897 | VCP/p97 | |
| 2 | View Details | [89..929] | 147.0 | Crystal Structure Analysis of ClpB |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.68 |
Source: Reynolds et al. (2008)