













| General Information: |
|
| Name(s) found: |
gi|6682244
[NCBI NR]
gi|15232736 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 777 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
DNA replication initiation
[ISS]
DNA replication [IEA] |
| Molecular Function: |
nucleotide binding
[IEA]
DNA binding [IEA][ISS] ATP binding [IEA][ISS] DNA-dependent ATPase activity [ISS] nucleoside-triphosphatase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGFYGGGSMA SGRPESTGID SLGIGKILAV YIKDNENLAI DEDKLQLTAE LIRVFSASPG 60
61 RDIVSQVNED GGGSFSLSLD LQQFKKISDI ENFFINLEDN PKGVIPCMNA AVHKVLFDQW 120
121 ETNEFENVMK INVRLHNYPE SSISLKNLRA AYIGKLVTVH GTVVKVSTVK PLVTQMAFDC 180
181 GKCKTGITRE FTDGKFSPPL KCDSHGCKSK TFTPIRSSAQ TIDFQKIRVQ ELQKPEDHEE 240
241 GRVPRTVECE LMEDLVDICI PGDVVTVTGI IGVINNYMDI GGGKSKTKNQ GFYYLFIEAV 300
301 SVKNTKRQSA FENSEDSSSS AQTADVGDLY SFSQRDLEFI VKFKEEYGSD TFRRILHSVC 360
361 PSIYGHEIVK AGITLSLFGG VRKHSMDRNK VPVRGDIHVI IVGDPGLGKS QLLQAAAAIS 420
421 PRGIYVCGNA TTRAGLTVAV VKDSMTNDYA FEAGAMVLAD GGLCCIDEFD KMTTEHQALL 480
481 EAMEQQCVSV AKAGLVASLS ARTSVIAAAN PVGGHYNRAK TVNENLKMSA ALLSRFDLVF 540
541 ILLDKPDELL DKQVSEHIMS HHRMLGMQTC MQKGILYFQD CGWTLRKMTT FLRFLANCLG 600
601 NIFPMHGILM SKDAGEIIQK FYLKLRDHNT SADSTPITAR QLESLVRLAQ ARARVDLREE 660
661 ITVQDAMDVV EIMKESLYDK LIDEHGVVDF GRSGGMSQQK EAKRFLSALD KQSELQQKDC 720
721 FSVSEMYSLA DRIGLRVPDI DTFLENLNIA GYLLKKGPKT YQVLSSSYSR SQSSRSR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
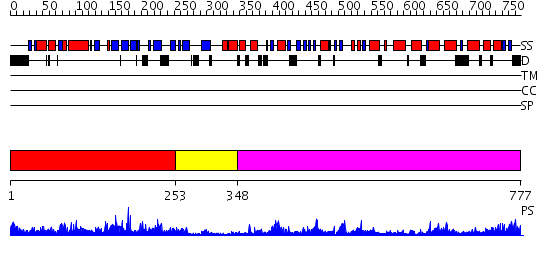
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..252] | 39.69897 | DNA replication initiator (cdc21/cdc54) N-terminal domain | |
| 2 | View Details | [253..347] | 37.39794 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 3 | View Details | [348..777] | 27.69897 | EDTA treated |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)