













| General Information: |
|
| Name(s) found: |
DHBF_BACSU
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Bacillus subtilis |
| Length: | 2378 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPDTKDLQYS LTGAQTGIWF AQQLDPDNPI YNTAEYIEIN GPVNIALFEE ALRHVIKEAE 60
61 SLHVRFGENM DGPWQMINPS PDVQLHVIDV SSEPDPEKTA LNWMKADLAK PVDLGYAPLF 120
121 NEALFIAGPD RFFWYQRIHH IAIDGFGFSL IAQRVASTYT ALIKGQTAKS RSFGSLQAIL 180
181 EEDTDYRGSE QYEKDRQFWL DRFADAPEVV SLADRAPRTS NSFLRHTAYL PPSDVNALKE 240
241 AARYFSGSWH EVMIAVSAVY VHRMTGSEDV VLGLPMMGRI GSASLNVPAM VMNLLPLRLT 300
301 VSSSMSFSEL IQQISREIRS IRRHHKYRHE ELRRDLKLIG ENHRLFGPQI NLMPFDYGLD 360
361 FAGVRGTTHN LSAGPVDDLS INVYDRTDGS GLRIDVDANP EVYSESDIKL HQQRILQLLQ 420
421 TASAGEDMLI GQMELLLPEE KEKVISKWNE TAKSEKLVSL QDMFEKQAVL TPERIALMCD 480
481 DIQVNYRKLN EEANRLARLL IEKGIGPEQF VALALPRSPE MVASMLGVLK TGAAYLPLDP 540
541 EFPADRISYM LEDAKPSCII TTEEIAASLP DDLAVPELVL DQAVTQEIIK RYSPENQDVS 600
601 VSLDHPAYII YTSGSTGRPK GVVVTQKSLS NFLLSMQEAF SLGEEDRLLA VTTVAFDISA 660
661 LELYLPLISG AQIVIAKKET IREPQALAQM IENFDINIMQ ATPTLWHALV TSEPEKLRGL 720
721 RVLVGGEALP SGLLQELQDL HCSVTNLYGP TETTIWSAAA FLEEGLKGVP PIGKPIWNTQ 780
781 VYVLDNGLQP VPPGVVGELY IAGTGLARGY FHRPDLTAER FVADPYGPPG TRMYRTGDQA 840
841 RWRADGSLDY IGRADHQIKI RGFRIELGEI DAVLANHPHI EQAAVVVRED QPGDKRLAAY 900
901 VVADAAIDTA ELRRYMGASL PDYMVPSAFV EMDELPLTPN GKLDRKALPA PDFSTSVSDR 960
961 APRTPQEEIL CDLFAEVLGL ARVGIDDSFF ELGGHSLLAA RLMSRIREVM GAELGIAKLF1020
1021 DEPTVAGLAA HLDLAQSACP ALQRAERPEK IPLSFAQRRL WFLHCLEGPS PTYNIPVAVR1080
1081 LSGELDQGLL KAALYDLVCR HESLRTIFPE SQGTSYQHIL DADRACPELH VTEIAEKELS1140
1141 DRLAEAVRYS FDLAAEPAFR AELFVIGPDE YVLLLLVHHI VGDGWSLTPL TRDLGTAYAA1200
1201 RCHGRSPEWA PLAVQYADYA LWQQELLGNE DDPNSLIAGQ LAFWKETLKN LPDQLELPTD1260
1261 YSRPAEPSHD GDTIHFRIEP EFHKRLQELA RANRVSLFMV LQSGLAALLT RLGAGTDIPI1320
1321 GSPIAGRNDD ALGDLVGLFI NTLVLRTDTS GDPSFRELLD RVREVNLAAY DNQDLPFERL1380
1381 VEVLNPARSR ATHPLFQIML AFQNTPDAEL HLPDMESSLR INSVGSAKFD LTLEISEDRL1440
1441 ADGTPNGMEG LLEYSTDLFK RETAQALADR LMRLLEAAES DPDEQIGNLD ILAPEEHSSM1500
1501 VTDWQSVSEK IPHACLPEQF EKQAALRPDA IAVVYENQEL SYAELNERAN RLARMMISEG1560
1561 VGPEQFVALA LPRSLEMAVG LLAVLKAGAA YLPLDPDYPA DRIAFMLKDA QPAFIMTNTK1620
1621 AANHIPPVEN VPKIVLDDPE LAEKLNTYPA GNPKNKDRTQ PLSPLNTAYV IYTSGSTGVP1680
1681 KGVMIPHQNV TRLFAATEHW FRFSSGDIWT MFHSYAFDFS VWEIWGPLLH GGRLVIVPHH1740
1741 VSRSPEAFLR LLVKEGVTVL NQTPSAFYQF MQAEREQPDL GQALSLRYVI FGGEALELSR1800
1801 LEDWYNRHPE NRPQLINMYG ITETTVHVSY IELDRSMAAL RANSLIGCGI PDLGVYVLDE1860
1861 RLQPVPPGVA GELYVSGAGL ARGYLGRPGL TSERFIADPF GPPGTRMYRT GDVARLRADG1920
1921 SLDYVGRADH QVKIRGFRIE LGEIEAALVQ HPQLEDAAVI VREDQPGDKR LAAYVIPSEE1980
1981 TFDTAELRRY AAERLPDYMV PAAFVTMKEL PLTPNGKLDR KALPAPDFAA AVTGRGPRTP2040
2041 QEEILCDLFM EVLHLPRVGI DDRFFDLGGH SLLAVQLMSR IREALGVELS IGNLFEAPTV2100
2101 AGLAERLEMG SSQSALDVLL PLRTSGDKPP LFCVHPAGGL GWCYAGLMTN IGTDYPIYGL2160
2161 QARGIGQREE LPKTLDDMAA DYIKQIRTVQ PKGPYHLLGW SLGGNVVQAM ATQLQNQGEE2220
2221 VSLLVMLDAY PNHFLPIKEA PDDEEALIAL LALGGYDPDS LGEKPLDFEA AIEILRRDGS2280
2281 ALASLDETVI LNLKNTYVNS VGILGSYKPK TFRGNVLFFR STIIPEWFDP IEPDSWKPYI2340
2341 NGQIEQIDID CRHKDLCQPE PLAQIGKVLA VKLEELNK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
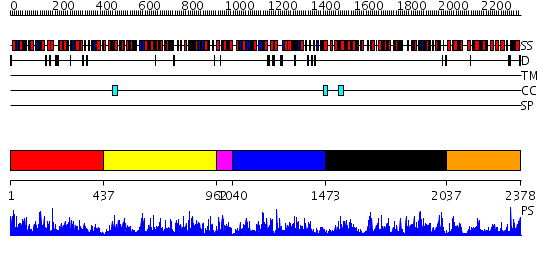
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..436] | 41.522879 | VibH | |
| 2 | View Details | [437..961] | 143.0 | Phenylalanine activating domain of gramicidin synthetase 1 | |
| 3 | View Details | [962..1039] | 16.69897 | No description for 2gdwA was found. | |
| 4 | View Details | [1040..1472] | 8.28 | VibH | |
| 5 | View Details | [1473..2036] | 112.0 | No description for 2p2bA was found. | |
| 6 | View Details | [2037..2378] | 7.522879 | Erythromycin polyketide synthase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)