













| General Information: |
|
| Name(s) found: |
trxr-1 /
CE34250
[WormBase]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 667 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
electron transport
[IEA removal of superoxide radicals [IEA glutathione metabolic process [IEA cell redox homeostasis [IEA detoxification of mercury ion [IEA |
| Molecular Function: |
disulfide oxidoreductase activity
[IEA NADP or NADPH binding [IEA oxidoreductase activity [IEA FAD binding [IEA mercury ion binding [IEA mercury (II) reductase activity [IEA thioredoxin-disulfide reductase activity [IEA glutathione-disulfide reductase activity [IEA dihydrolipoyl dehydrogenase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSLTELFGC FKRQPSQQEA SSPANPHVSD TLSMGVAASG MPPPKRPAPA ESPTLPGETL 60
61 VDAPGIPLKE ALKEAANSKI VIFYNSSDEE KQLVEFETYL NSLKEPADAE KPLEIPEMKK 120
121 LQVSRASQKV IQYLTLHTSW PLMYIKGNAV GGLKELKALK QDYLKEWLRD HTYDLIVIGG 180
181 GSGGLAAAKE ASRLGKKVAC LDFVKPSPQG TSWGLGGTCV NVGCIPKKLM HQASLLGHSI 240
241 HDAKKYGWKL PEGKVEHQWN HLRDSVQDHI ASLNWGYRVQ LSEKTVTYIN SYGEFTGPFE 300
301 ISATNKKKKV EKLTADRFLI STGLRPKYPE IPGVKEYTMT SDDLFQLPYS PGKTLCVGAS 360
361 YVSLECAGFL HGFGFDVTVM VRSILLRGFD QDMAERIRKH MIAYGMKFEA GVPTSIEQID 420
421 EKTDEKAGKY RVFWPKKNEE TGEMQEVSEE YNTILMAIGS EAVTDDVGLT TIGVESAKSK 480
481 KVLGSREQST TIPWVYAIGD VLEGTPELTP VAIQAGSVLM SRIFDGANEL TEYDQIPTTV 540
541 FTPLEYGCCG LSEEDAMMKY GKDNIMIYHN VFNPLEYTIS ESMDKDHCYL KMICLSNEEE 600
601 KVVGFHILTP NAGEVTQGFG IALKLAAKKA DFDRLIGIHP TVAENFTTLT LEKKEGDEEL 660
661 QASGCUG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
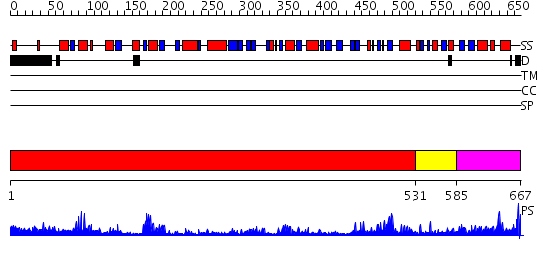
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..530] | 55.522879 | Alkyl hydroperoxide reductase subunit F (AhpF), C-terminal domains; Alkyl hydroperoxide reductase subunit F (AhpF), N-terminal domain | |
| 2 | View Details | [531..584] | 29.522879 | NADH oxidase /nitrite reductase from Pyrococcus furiosus Pfu-1140779-001 | |
| 3 | View Details | [585..667] | 92.30103 | Mammalian thioredoxin reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)