













| General Information: |
|
| Name(s) found: |
pef-1 /
CE27999
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 707 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA cytoplasm [IEA neuronal cell body [IDA |
| Biological Process: |
detection of stimulus involved in sensory perception
[IEA protein amino acid dephosphorylation [IEA |
| Molecular Function: |
phosphoprotein phosphatase activity
[IEA bis(5'-nucleosyl)-tetraphosphatase (symmetrical) activity [IEA iron ion binding [IEA calcium ion binding [IEA manganese ion binding [IEA hydrolase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGCGPSSGRQ NPSTELKKST RATTTTTSSS QRNNYNDNNQ NTSSSSGNKK ESSSSSKQHS 60
61 SKKSKKSNSK KNRSPSPQPQ LTIKSAILIQ KWYRRCEARL EARRRATWQI FTALEYAGEQ 120
121 DQLKLYDFFA DVIRAMAEEN GKGGVENGRN SPLMSALSHY AKPSLMDSEG ETVKKMLEDT 180
181 SPTNVDIDRN YKGPTLSLPL DKPQVAKMIE AFKVNKVLHP KYVLMILHEA RKIFKAMPSV 240
241 SRISTSISNQ VTICGDLHGK FDDLCIILYK NGYPSVDNPY IFNGDFVDRG GQSIEVLCVL 300
301 FALVIVDPMS IYLNRGNHED HIMNLRYGFI KELSTKYKDL STPITRLLED VFSWLPIATI 360
361 IDRDIFVVHG GISDQTEVSK LDKIPRHRFQ SVLRPPVNKG MESEKENSAV NVDEWKQMLD 420
421 IMWSDPKQNK GCWPNVFRGG GSYFGADITA SFLEKHGFRL LVRSHECKFE GYEFSHNNTC 480
481 LTVFSASNYY ETGSNRGAYV KFIGKSKQPH FVQYMASKTH RKSTLRERLG VVEESAVKEL 540
541 KEKLSSFHTD LQKEFEIMDI EKSGKLPILK WSDCVERITG LNLPWIALAP KVATLSEDGK 600
601 YVMYKEDRRI AQVGGTHAQE KDIVESLYRH KSTLETLFRF MDKDNSGQVS MKEFIDACEV 660
661 LGKYTKRPLQ TDYISQIAES IDFNKDGFID LNELLEAFRL VDRPLLR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
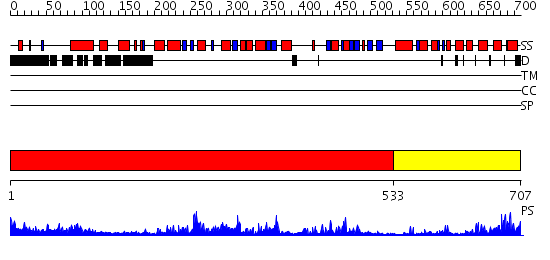
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..532] | 104.0 | PP5 structure | |
| 2 | View Details | [533..707] | 30.0 | Structure of calmodulin bound to a calcineurin peptide: a new way of making an old binding mode |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)