













| General Information: |
|
| Name(s) found: |
rad-51 /
CE25285
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 357 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA germ cell nucleus [IDA intracellular [IEA condensed nuclear chromosome [IDA |
| Biological Process: |
DNA repair
[IEA meiotic chromosome condensation [IMP reciprocal meiotic recombination [IEA embryonic development ending in birth or egg hatching [IMP negative regulation of apoptosis [IMP DNA recombination [IEA DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis [IGI chromosome segregation [IMP response to DNA damage stimulus [IMP DNA metabolic process [IEA |
| Molecular Function: |
nucleotide binding
[IEA DNA binding [IEA ATP binding [IEA DNA-dependent ATPase activity [IEA nucleoside-triphosphatase activity [IEA sequence-specific DNA binding [IEA damaged DNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAQASRQKK SDQEQRAADQ ALLNAAIEDN AMEQDENFTV IDKLESSGIS SGDISKLKEA 60
61 GYYTYESLAF TTRRELRNVK GISDQKAEKI MKEAMKFVQM GFTTGAEVHV KRSQLVQIRT 120
121 GSASLDRLLG GGIETGSITE VYGEYRTGKT QLCHSLAVLC QLPIDMGGGE GKCMYIDTNA 180
181 TFRPERIIAI AQRYNMDSAH VLENIAVARA YNSEHLMALI IRAGAMMSES RYAVVIVDCA 240
241 TAHFRNEYTG RGDLAERQMK LSAFLKCLAK LADEYGVAVI ITNQVVAQVD GGASMFQADA 300
301 KKPIGGHIIA HMSTTRLYLR KGKGENRVAK MVQSPNLPEA EATYSITNHG IEDARED |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
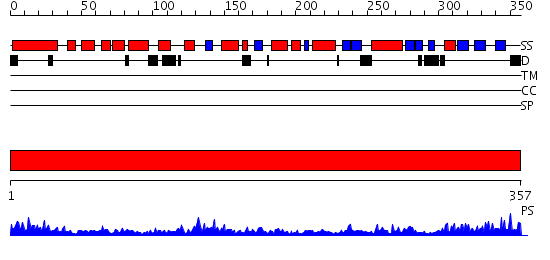
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..357] | 77.522879 | A Crystal Structure of the Rad51 Filament |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)