













| General Information: |
|
| Name(s) found: |
klp-16 /
CE08666
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 587 amino acids |
Gene Ontology: |
|
| Cellular Component: |
microtubule associated complex
[IEA |
| Biological Process: |
embryonic development
[IMP embryonic cleavage [IMP embryonic development ending in birth or egg hatching [IMP chromosome segregation [IMP cytokinesis [IMP microtubule-based movement [IEA meiosis [IMP |
| Molecular Function: |
ATP binding
[IEA microtubule motor activity [IEA motor activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNVARRRSGL FRSTIGAPPK ATRGRAAAPP IKEADPATIP RQSAPGGITI GAAACRPPSR 60
61 LPGATISATG RASLPERSAM AKTSTSSHSR PALQSTASRN TTLTAASTSR QLRTGRPPPP 120
121 STQRSTATFS LKPSVARARP VAQKPILPSK VTLLEERNAQ LEREMAEATD FAEHQKSKIQ 180
181 FLDGKLEGAD RKLISLQDQL STLKEVHKSK MEECEEYRVH NNDLRDLLNE KEAELRKLHN 240
241 DVVDLRGQIR VAVRVRPLIK SEVDTSSSAI EYPAVDAIKI NEGSKPGNVV KFENVFTPVF 300
301 SQKEVFANVE EFIRSSLHGY NVGLIAYGQT GSGKTHTMRG GNGEEEGIIP RAAAFLFAES 360
361 KKLESLGWKF DFSLSFLEVY NNVAYDLLSG RDVVQLRLND QTVSMIGLSE HTISNVSDVA 420
421 RLLRVADGGR KTAATKCNGS SSRSHAVYMW KIKAHQPSTG ISTTCQLKLV DLAGSERAKE 480
481 SGVSGDQFKE MTNINQSLSI LQMCISQQRS QKGHVSYRDS KLTQVLMDCL GRGSSKTMVV 540
541 VNLNPCNEQA TESKRSIEFA SKMRSTHIGS AVQQRTLLGD VSQMSMN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
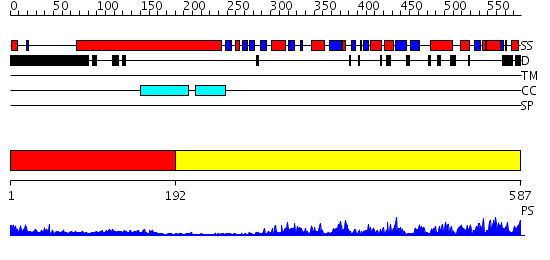
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | 10.221849 | Heavy meromyosin subfragment | |
| 2 | View Details | [192..587] | 66.39794 | Kinesin motor Ncd (non-claret disjunctional) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)