













| General Information: |
|
| Name(s) found: |
tea2 /
SPBC1604.20c
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 628 amino acids |
Gene Ontology: |
|
| Cellular Component: |
microtubule cytoskeleton
[IDA cytoplasm [IDA cell cortex of cell tip [IDA |
| Biological Process: |
regulation of filamentous growth
[IMP establishment or maintenance of cell polarity [IMP microtubule-based movement [ISS cellular protein localization [IGI |
| Molecular Function: |
ATPase activity
[IDA ATP binding [IEA] protein binding [IPI protein heterodimerization activity [IPI plus-end-directed microtubule motor activity [IDA microtubule binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSSSKPVN TGLVTPRRYS TMTGIRTGPS QSGTGSIPYS PTSPLSRNFS NYSIPMLRSN 60
61 STQTNVNGPT AFDLGVTEKL MSPGTLDRYT RPALYPSKDL DYLKNEYVNY ESTTSQQTNS 120
121 KGLKESNFVG SGIITSIRIR PIGKNQGVWS HGKLSNDPYG REYIRQQTST SSSTIQQEYL 180
181 FNNVFGMESK NYDIYKRSVK SVVRNVFSGY NGIVFAYGMT GTGKTYSMQG TENEPGIIPL 240
241 AMNDLFEMVE NNSDDDTFQI RISYLEIYNE RIRDLIGNSD EEPRIRENAS GEVNVTPLTR 300
301 VLVTSPEEVS QVIEQCNAIR KTAATDFNTY SSRSHAILQV FLIRNNPTAH TSQISSLSLV 360
361 DLAGSERASA HHERRKEGAF INKSLLTLGT VISRLSAAAN PSLTSNSGHI PYRESKLTRL 420
421 LQQSLSGQSQ ISLLATISIE SNHTMETTNT LKFASRAQNL PQDIRQAEAV TNVQAELASL 480
481 HSALEKNAQE VEYYASLVKQ LTSDLEERDT YIAMLEAERS QGTAISRARL RMEELLSDHN 540
541 FEIADLRDEL QDKEQIIYAL RYAQKQRDIA DFNQSLAKFP HKILKKNVTR GSRSSSDQFS 600
601 NETKTEILPD DQQQSKKDSV TQETQLLS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
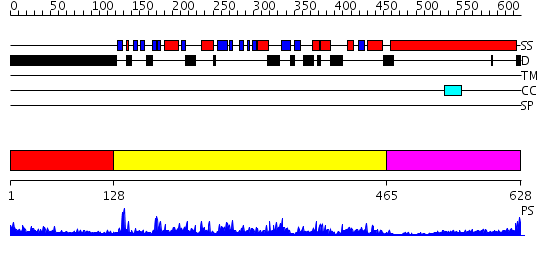
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..127] | 3.703996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [128..464] | 96.221849 | Kinesin | |
| 3 | View Details | [465..628] | 4.30103 | No description for 2dfsA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)