













| General Information: |
|
| Name(s) found: |
SPBC1685.08
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 424 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Rpd3L-Expanded complex [IDA cytoplasm [IDA Rpd3S complex [IDA |
| Biological Process: |
chromatin remodeling
[NAS]
regulation of transcription, DNA-dependent [ISS histone deacetylation [ISS gene-specific transcription from RNA polymerase II promoter [NAS] |
| Molecular Function: |
zinc ion binding
[IEA]
transcription factor binding [ISS small conjugating protein ligase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSNSTPANE EKEEESEKIQ KSPVDTPHTP NGSVSDNEEN ETSSTGEVTR CVCGIVESDD 60
61 EASDGGLYIQ CDQCSVWQHG NCVGFADESE VPEVYYCEIC HPEFHKVYQR GRGAKQSKYL 120
121 GNGKPIEASQ TEESSSTPPS PATKKSSKQR LTMNSRDAAL DYEEYLAIAK EKSLIPRRSR 180
181 GRTSSKSLSP PAPQDESQGT EINLKQKIEE ENDEILEDSK ESKDENEENK ETSTTNVAET 240
241 DAPEEETVDT VEEIADEEKH SVKEESGEAS PQSSQQSTIT SISTTTRSTR KAKREAAAED 300
301 KADLPAAVAP KPSKTRKVGG RRGKSSSNDN HRIPQLHPDG TFVETITKPK GLHSRITMTE 360
361 MRRRVASMLE YIGHIQVEMA AQSAGNQSST KSSKEGPEEE KETLRMVDNL TRDLLHWEQR 420
421 FSRT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
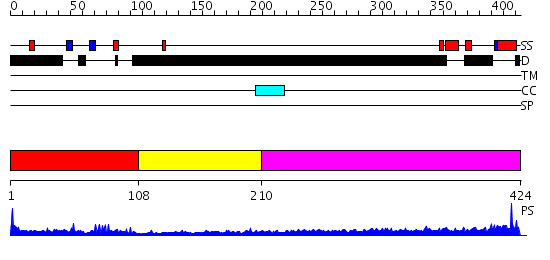
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | 8.69897 | Solution structure of PHD domain in death inducer-obliterator 1(DIO-1) | |
| 2 | View Details | [108..209] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [210..424] | 1.172996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)