













| General Information: |
|
| Name(s) found: |
gnr1 /
SPAC343.04c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 507 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [ISS spindle pole body [IDA cytosol [IDA GID complex [ISS] |
| Biological Process: |
proteasomal ubiquitin-dependent protein catabolic process
[ISS]
negative regulation of gluconeogenesis [ISS] |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALDEKFQLE VIHLLLQFLN DYGYDESLKA LEKETGLVSE TEDVKRLKQA VLQGDWITAE 60
61 AAFSIMQLRD ESKRKEAQFL LQKQRCLELA RSGAICEAIY VLQNFESTDF NKEKERLVSI 120
121 ILESNNKSNN ELITKNGYGN TRLDLLNQLS EYISPEILLP KRRLEHLLQQ AKDYQVSSQV 180
181 YHNVLKNFSF LSDYKADPSE LPTKEYHVFH DHSDEVWQIS YSHNGRYLAS ASKDKTAIIF 240
241 DVVNLKRVFR LIGHIDTVAY IRWSPDDRYL LSCSCDKSVI LWDAFTGEKL RDYKHGFSVS 300
301 CCCWLPDGLS FITGSPDCHI THWSLNGEIL YKWEDVNIYD MALTSDGTKL YIVGFEQLIN 360
361 AEDKHIAIYS VETRECIKKI SLQSKVTSIC LSKDSKYALT NLEPHTTFLW DLEENRIVRQ 420
421 YMGHKLGNFL IGSCFGGKDD TFVLSGSEDD KIRIWHRESG KLLATLSGHV KCVNYVAYNP 480
481 VDPYQFASAG DDNTVRIWSN KDNPRRQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
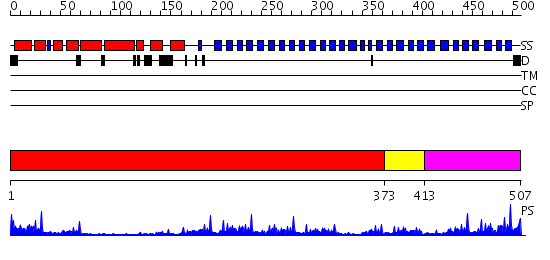
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..372] | 13.221849 | Crystal Structure analysis of the beta-propeller protein Ski8p | |
| 2 | View Details | [373..412] | 32.69897 | Groucho/tle1, C-terminal domain | |
| 3 | View Details | [413..507] | 62.69897 | No description for 2gnqA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)