













| General Information: |
|
| Name(s) found: |
SPAC186.09
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 572 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
cellular amino acid catabolic process
[ISS]
generation of precursor metabolites and energy [NAS] pyruvate metabolic process [ISS |
| Molecular Function: |
magnesium ion binding
[IEA]
thiamin pyrophosphate binding [IEA] pyruvate decarboxylase activity [ISS transferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKDNQQAVQL QQPTTIGHYL AVRLAQAGVK HHFVVPGDYN LGLLDKLQYN NYLEEVNCAN 60
61 ELNCAFAAEG YARANGIAAC VVTYSVGAFT AFDGIGGAYA EDLPVILISG SPNTNDIGSS 120
121 HLLHHTLGTH DFSYQYEMAK KITCAAVSIQ RPTEAPRLID YAIKMALLKK KPVYIEVPTN 180
181 VASQPCAAPG PASLITEPET SNQEYLQMAV DISAKIVNGK QKPVLLAGPK LRSFKAESAF 240
241 LELANSLNCS VAVMPNAKSF FPESHPNYAG IYWGQASTLG AESIINWSDC IICAGTTFTD 300
301 YSSNGWTSLP PKANVLHVDV DRVTVSDAEF GGVLLRDFLH ELAKKVKANN ASVVEYKRIR 360
361 PESLEIPMEN PKAALNRKEI IRQVQNLVNQ ETTLFVDTGD SWFGGMRITL PEKARFEIEM 420
421 QWGHIGWSVP SAFGYAIGAP KRNVVVFVGD GSFQETVQEV SQMVRLNLPI IMFLINNRGY 480
481 TIEVEIHDGP YNRIKNWDYA AIVEAFNAGE GHAKGFRVGN GHELAEAIRQ AKENSQGPTL 540
541 IECNIDQDDC SKELINWGHN VGAANGKPPA KE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
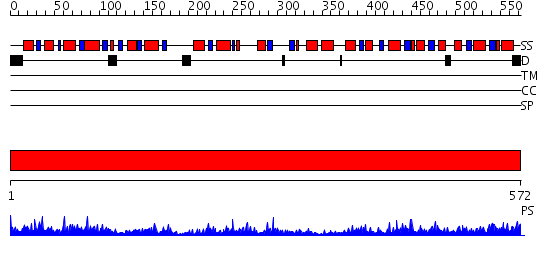
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..572] | 96.69897 | Pyruvate decarboxylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)