













| General Information: |
|
| Name(s) found: |
SPAC3G6.11
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 844 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC cytosol [IDA |
| Biological Process: |
mitotic sister chromatid segregation
[ISS DNA recombination [NAS chromatin silencing [NAS |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] ATP-dependent DNA helicase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCHSKEVKFK TNFHHPYTPY DIQLEFMRSL YSSISDGKIG IFESPTGTGK SLSLICASLT 60
61 WLDEHGGVLL EDNEKSNDNK SNTSSKIPDW VLEQDLKIQK DLVKETHARL EQRLEEIRKR 120
121 NQSRKNQMSN NSTTYHRETK RRNINAEAST SDNCNNSNTS VDPMDEYLVT AEYTMPSTSE 180
181 QSEDLFNNGY SSKVSELLRK LSPDNEKPPI VQKIYFTSRT HSQLQQLVQE IKKLNNQTFS 240
241 TPIRVVSLAS RKNLCINNEV RKLRPTSALN EKCIELQGSA HKCPFLQDNT QLWDFRDEAL 300
301 AEIMDIEELV ELGQRLKVCP YYGTREAVDS AQIVTLPYPL LLQESARNAL NLTLKDNICI 360
361 IDEAHNLIDA ICSMHSSSIS FRQVCIAETQ LQQYFLRFEK RLNGNNRMHI KQLIKVVYNL 420
421 KSFFLNCLET NTNSKVINVD SLLVSNGADQ INLHHLSEYL NVSKLARKVD GYTKYMHSLG 480
481 TQELESLNDL RSERFSNGNG YEEDPYTPVL MQLESFLLNI ANPAPEGKLF YEKQTGDNPY 540
541 LKYLLLDPSK HVEILTEQCR SVNLAGGTMS PIDDFITLLF SDEQSRILPF SCDHIVPPEN 600
601 ITTILVSQGP AGVPFEFTHK RKDDENLLKD LGRTFQNFIS IIPDGVVVFF PSFAFLQQAV 660
661 KVWEMNGITN RLNAKKPLFI ESKDFGDNPL DTFEHYKQSV DAGLSGMLFS VIGGRLSEGI 720
721 NFSDKLGRAV MVVGMPFPNS QDVEWQAKVS YVEEKAKEKG INAKQASQEF YENTCMRAVN 780
781 QSIGRAIRHR DDYASIILLD SRYNRSSIQR KLPNWLSKNI HSSPNFGPAI RQLATFFRAK 840
841 KMCD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
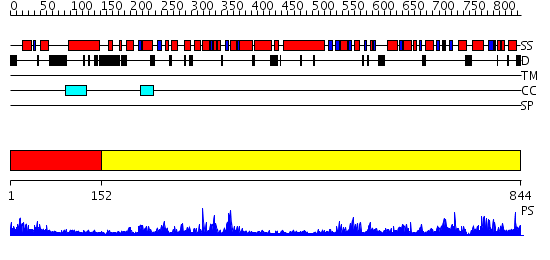
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 2.204989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [152..844] | 1.17 | No description for 2p6rA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)