













| General Information: |
|
| Name(s) found: |
ksg1 /
SPCC576.15c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 592 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
ascospore formation
[IMP signal transduction [IC] cellular cell wall organization [IMP protein amino acid phosphorylation [IEA] regulation of conjugation with cellular fusion [IMP |
| Molecular Function: |
ATP binding
[IEA]
protein binding [IPI 3-phosphoinositide-dependent protein kinase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRNTHNPNET EASEDAENDT QSESDLSFDH GSSEKLNRAS LPKTQNSAIP QSNALNTTPN 60
61 ESTSQIDSSP KIPSAVPHIS TPNPSSGAST PNIKRVSDFK FGEILGEGSY STVLTATENS 120
121 TKREYAIKVL DKRHIIKEKK EKYVNIEKEA LCILSKHPGF IKLFYTFQDA HNLYFVLSLA 180
181 RNGELLDYIN KLGRFNEICA QYYAALIVDS IDYMHGRGVI HRDLKPENIL LDDNMRTKIT 240
241 DFGSAKILNS SHGSHEEDTH HADKPQAHSR SFVGTARYVS PEVLSDKIAG TASDIWAFGC 300
301 ILFQMLAGKP PFVAGNEYLT FQSILHLSYE IPPDISDVAS DLIKKLLVLD PKDRLTVDEI 360
361 HQHPFFNGIK FDNTLWELPP PRLKPFGHTS VLSLSVPNAS NKHENGDLTS PLGVPSMVSA 420
421 STNAAPSPVG TFNRGTLLPC QSNLEEENKE WSSILQDDEK ISKIGTLNVY SMSGINGNDA 480
481 FRFFSSLFRK RKPRTFILTN FGRYLCVASD GEGRKTVKEE IPIKSVGMRC RMVKNNEHGW 540
541 VVETPTKSWS FEDPNGPASA WVELLDKASS ISLPFGNHSV TSFSRSIARS AV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
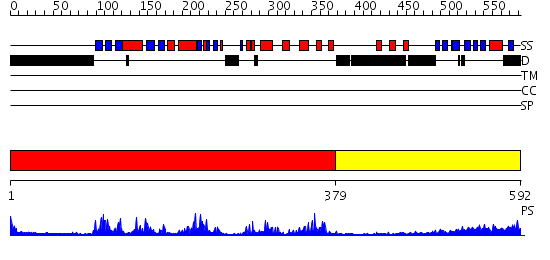
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..378] | 65.522879 | Structure of human PDK1 kinase domain in complex with staurosporine | |
| 2 | View Details | [379..592] | 15.69897 | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)