













| General Information: |
|
| Name(s) found: |
psm3 /
SPAC10F6.09c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1194 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC cytosol [IDA mitotic cohesin complex [TAS condensed nuclear chromosome [IC |
| Biological Process: |
mitotic sister chromatid cohesion
[IMP protein ubiquitination [ISS |
| Molecular Function: |
ATPase activity
[ISS ATP binding [IEA] protein binding [IPI protein heterodimerization activity [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYITKIVIQG FKSYKDYTVI EPLSPHHNVI VGRNGSGKSN FFAAIRFVLS DAYTHLSREE 60
61 RQALLHEGPG ATVMSAYVEV TFANADNRFP TGKSEVVLRR TIGLKKDEYS LDKKTVSKTE 120
121 VINLLESAGF SRSNPYYIVP QGRVTSLTNA KDSERLELLK EVAGTQIYEN RRAESNKIMD 180
181 ETIQKSEKID ELLQYIEERL RELEEEKNDL AVYHKKDNER RCLEYAIYSR EHDEINSVLD 240
241 ALEQDRIAAL ERNDDDSGAF IQREERIERI KAEITELNHS LELLRVEKQQ NDEDYTNIMK 300
301 SKVALELQSS QLSRQIEFSK KDESSKLNIL SELESKISEK ENELSEILPK YNAIVSEADD 360
361 LNKRIMLLKN QKQSLLDKQS RTSQFTTKKE RDEWIRNQLL QINRNINSTK ENSDYLKTEY 420
421 DEMENELKAK LSRKKEIEIS LESQGDRMSQ LLANITSINE RKENLTDKRK SLWREEAKLK 480
481 SSIENVKDDL SRSEKALGTT MDRNTSNGIR AVKDIAERLK LEGYYGPLCE LFKVDNRFKV 540
541 AVEATAGNSL FHIVVDNDET ATQILDVIYK ENAGRVTFMP LNKLRPKAVT YPDASDALPL 600
601 IQYLEFDPKF DAAIKQVFSK TIVCPSIETA SQYARSHQLN GITLSGDRSD KKGALTAGYR 660
661 DYRNSRLDAI KNVKTYQIKF SDLQESLEKC RSEIESFDQK ITACLDDLQK AQLSLKQFER 720
721 DHIPLKDELV TITGETTDLQ ESMHHKSRML ELVVLELHTL EQQANDLKSE LSSEMDELDP 780
781 KDVEALKSLS GQIENLSHEF DAIIKERAHI EARKTALEYE LNTNLYLRRN PLKAEIGSDN 840
841 RIDESELNSV KRSLLKYENK LQIIKSSSSG LEEQMQRINS EISDKRNELE SLEELQHEVA 900
901 TRIEQDAKIN ERNAAKRSLL LARKKECNEK IKSLGVLPEE AFIKYVSTSS NAIVKKLHKI 960
961 NEALKDYGSV NKKAYEQFNN FTKQRDSLLA RREELRRSQE SISELTTVLD QRKDEAIERT1020
1021 FKQVAKSFSE IFVKLVPAGR GELVMNRRSE LSQSIEQDIS MDIDTPSQKS SIDNYTGISI1080
1081 RVSFNSKDDE QLNINQLSGG QKSLCALTLI FAIQRCDPAP FNILDECDAN LDAQYRSAIA1140
1141 AMVKEMSKTS QFICTTFRPE MVKVADNFYG VMFNHKVSTV ESISKEEAMA FVEG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
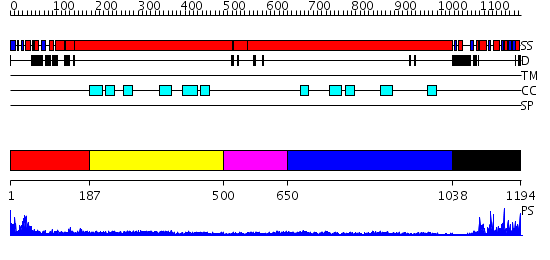
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..186] | 37.154902 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. | |
| 2 | View Details | [187..499] | 2.9 | Tropomyosin | |
| 3 | View Details | [500..649] | 5.13 | Smc hinge domain | |
| 4 | View Details | [650..1037] | 2.07 | The modeled structure of fibritin (gpwac) of bacteriophage T4 based on cryo-EM reconstruction of the extended tail of bacteriophage T4 | |
| 5 | View Details | [1038..1194] | 18.522879 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)